

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.5 + phase: 0
(377 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
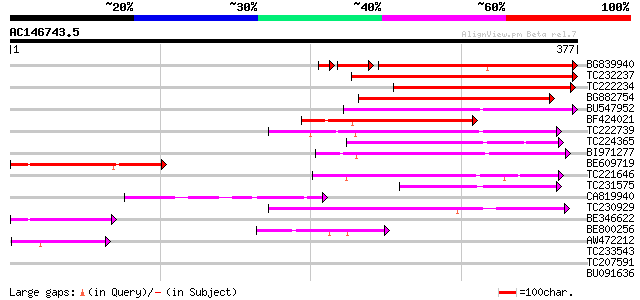
Score E
Sequences producing significant alignments: (bits) Value
BG839940 227 4e-66
TC232237 166 2e-41
TC222234 132 3e-31
BG882754 124 7e-29
BU547952 107 1e-23
BF424021 101 5e-22
TC222739 94 1e-19
TC224365 91 9e-19
BI971277 80 1e-15
BE609719 78 7e-15
TC221646 71 7e-13
TC231575 70 1e-12
CA819940 65 4e-11
TC230929 weakly similar to UP|Q6ZCP9 (Q6ZCP9) Pentatricopeptide ... 60 1e-09
BE346622 47 1e-05
BE800256 43 3e-04
AW472212 42 5e-04
TC233543 39 0.003
TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E608... 30 2.3
BU091636 similar to GP|6730969|dbj beta-amyrin synthase {Glycyrr... 29 3.0
>BG839940
Length = 635
Score = 227 bits (578), Expect(3) = 4e-66
Identities = 111/134 (82%), Positives = 118/134 (87%), Gaps = 2/134 (1%)
Frame = +2
Query: 246 LRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINN 305
+RN VA EQ HYPFQTYI DY + FLIDTSQDVD LV+KGIMIN LGDS+AVA M+NN
Sbjct: 122 IRNXVAIEQGHYPFQTYIXDYIFVFXFLIDTSQDVDTLVDKGIMINXLGDSSAVANMVNN 301
Query: 306 LCLNVVQENIN--GGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLL 363
LCLNVVQENIN GGYI LCRKLNCFYEDPSHKYKAIF+HDYFSTPWKITSF AAIVLL
Sbjct: 302 LCLNVVQENININGGYIYLCRKLNCFYEDPSHKYKAIFMHDYFSTPWKITSFIAAIVLLF 481
Query: 364 LTLIQATCSIISLF 377
LTLIQATCS+ISLF
Sbjct: 482 LTLIQATCSVISLF 523
Score = 42.7 bits (99), Expect(3) = 4e-66
Identities = 17/24 (70%), Positives = 19/24 (78%)
Frame = +1
Query: 219 QSVLELKFAKGALTIPRFEVCHWT 242
+ LEL+F KG LT PRFEVCHWT
Sbjct: 40 KGALELQFEKGVLTXPRFEVCHWT 111
Score = 21.2 bits (43), Expect(3) = 4e-66
Identities = 9/11 (81%), Positives = 10/11 (90%)
Frame = +3
Query: 206 VEAGVKLHVGQ 216
VEAGVKL VG+
Sbjct: 3 VEAGVKLQVGK 35
>TC232237
Length = 673
Score = 166 bits (420), Expect = 2e-41
Identities = 86/151 (56%), Positives = 105/151 (68%), Gaps = 1/151 (0%)
Frame = +1
Query: 228 KGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQT-YITDYTILLDFLIDTSQDVDKLVEK 286
+G LT+P + +E RN+VAFEQCH T IT Y +LDFLI+T +DV+ LV+K
Sbjct: 43 EGVLTMPILNIADDSEMFFRNIVAFEQCHLSDDTNIITQYRKILDFLINTEKDVNVLVDK 222
Query: 287 GIMINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHDYF 346
I++N +GD+NAVA M+N+L N+ N Y SLC LN FYE P +KYKAIF HDYF
Sbjct: 223 KIIVNWMGDANAVATMVNSLGSNIGMPRFNPVYFSLCNSLNDFYESPCNKYKAIFKHDYF 402
Query: 347 STPWKITSFAAAIVLLLLTLIQATCSIISLF 377
+TPWKI S AAIVLLLLTLIQ CSI SLF
Sbjct: 403 NTPWKIASTVAAIVLLLLTLIQTICSINSLF 495
>TC222234
Length = 580
Score = 132 bits (332), Expect = 3e-31
Identities = 59/121 (48%), Positives = 84/121 (68%)
Frame = +3
Query: 256 HYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENI 315
HYP+ +++TDY +LDFL++TS+DVD LV KG+++N LGDS++VA M N L NV N
Sbjct: 3 HYPYHSFVTDYVAVLDFLVNTSRDVDVLVRKGVLVNWLGDSDSVADMFNGLWKNVTHINF 182
Query: 316 NGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSIIS 375
+ Y LC+KLN F +P HK K+ DY TPW+I A IVLL+L+L+Q+ CS++
Sbjct: 183 SSHYSELCQKLNAFCRNPCHKLKSTLRRDYCKTPWQIAVSIAGIVLLVLSLVQSVCSVLQ 362
Query: 376 L 376
+
Sbjct: 363 V 365
>BG882754
Length = 399
Score = 124 bits (311), Expect = 7e-29
Identities = 62/131 (47%), Positives = 87/131 (66%), Gaps = 1/131 (0%)
Frame = +2
Query: 233 IPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINT 292
IP+ V TETL RN+VA EQCHY FQ+YITDY LDFL++T++DVD LV++ + IN
Sbjct: 5 IPQLVVDDLTETLFRNMVALEQCHYLFQSYITDYVGFLDFLVNTNRDVDILVQERVFINW 184
Query: 293 LGDSNAVAKMINNLCLNVVQE-NINGGYISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWK 351
LGD+++VA MIN L N+ N + Y+ + KLN F+++P K K+ DY PW+
Sbjct: 185 LGDTDSVATMINGLMKNITTPINTSSQYLDVSEKLNAFHKNPWRKLKSALRRDYCRGPWQ 364
Query: 352 ITSFAAAIVLL 362
+ AA++LL
Sbjct: 365 TAASTAAVILL 397
>BU547952
Length = 699
Score = 107 bits (266), Expect = 1e-23
Identities = 63/156 (40%), Positives = 87/156 (55%), Gaps = 1/156 (0%)
Frame = -3
Query: 223 ELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDK 282
++KF G L IPR + T++L N++A EQCH IT Y I +D LI++ +DV
Sbjct: 670 DIKFKDGKLRIPRLLIHDGTKSLFLNLIAXEQCHLDCSNDITSYVIFMDNLINSPEDVGY 491
Query: 283 LVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGGYIS-LCRKLNCFYEDPSHKYKAIF 341
L +GI+ + LG VA + N LC VV + IN Y+S L +N +Y + + A
Sbjct: 490 LHYRGIIEHWLGSDAEVADLFNRLCQEVVFD-INNSYLSPLSEDVNRYYNHRWNTWCASL 314
Query: 342 IHDYFSTPWKITSFAAAIVLLLLTLIQATCSIISLF 377
H+YFS PW I S AA+VLLLLTL Q I +
Sbjct: 313 RHNYFSNPWAIISLVAAVVLLLLTLAQTYYGIYGYY 206
>BF424021
Length = 421
Score = 101 bits (252), Expect = 5e-22
Identities = 54/119 (45%), Positives = 78/119 (65%), Gaps = 2/119 (1%)
Frame = +3
Query: 195 IVKHLFSASQLVEAGVKLHVGQDYQSVLELKF--AKGALTIPRFEVCHWTETLLRNVVAF 252
+V HL SASQL E G+ + + + ELK+ KG + +P + TETL RN++A
Sbjct: 57 MVNHLPSASQLSEVGM-VFKASSCKCLFELKYHHRKGVMEMPCLTIEDRTETLFRNILAL 233
Query: 253 EQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVV 311
EQCHY +T + LL+FLI+T +DV+ LV+K I++N +GD+NAV KM N+LC NV+
Sbjct: 234 EQCHYILSPNVTQFLFLLNFLINTEKDVNILVDKKIIVNWMGDANAVVKMFNSLCSNVI 410
>TC222739
Length = 756
Score = 94.0 bits (232), Expect = 1e-19
Identities = 69/228 (30%), Positives = 106/228 (46%), Gaps = 33/228 (14%)
Frame = +2
Query: 173 NLTDLLRVFYLPPDMLPKREKQIVKH-------------LFSASQLVEAGVKLHVGQDYQ 219
+ TDL+R +LP + + + + L +A++L EAGV Q +
Sbjct: 38 HFTDLIRYTFLPTKIQVEGVNNVSVNPSQHFTPCQVECVLRTATKLNEAGVSFEKVQG-R 214
Query: 220 SVLELKFAK--------------------GALTIPRFEVCHWTETLLRNVVAFEQCHYPF 259
S L++KF K L IP +V TE +LRN++A EQCHY
Sbjct: 215 SYLDIKFEKTPILSWFLCFGCLPFSKWFKARLQIPHLKVDQVTECVLRNLIALEQCHYSD 394
Query: 260 QTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGGY 319
Q +I +Y L+D LI T +DV+ LV+ I+ + LG +A MIN LC +V+ + Y
Sbjct: 395 QPFICNYVTLIDSLIHTQEDVELLVDTEIIDHELGSHTELATMINGLCKHVLV--TSNYY 568
Query: 320 ISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLI 367
++LN Y Y + I YF PW+ +S I + L ++
Sbjct: 569 GKTTKELNEHYNCCWKHYMGMLISVYFRDPWRFSSTIVGIAVFLFAVV 712
>TC224365
Length = 766
Score = 90.9 bits (224), Expect = 9e-19
Identities = 53/145 (36%), Positives = 79/145 (53%), Gaps = 1/145 (0%)
Frame = +1
Query: 225 KFAKGALTIPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLV 284
K K L IP+ +V H TE + +N+VAFEQ HYP + Y +Y +D LI T DV+ LV
Sbjct: 91 KLFKARLRIPQLKVDHTTERVFKNLVAFEQFHYPDKPYFCNYVSFIDSLIHTQLDVELLV 270
Query: 285 EKGIMINTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFY-EDPSHKYKAIFIH 343
EK ++++ LG VA ++N LC +VV + Y + KLN Y D +H A+ +
Sbjct: 271 EKEVIVHELGSDKEVATLVNGLCKHVVTNST--CYHHIINKLNDHYMNDWNHTIAALRL- 441
Query: 344 DYFSTPWKITSFAAAIVLLLLTLIQ 368
YF W+ + I +L+ + Q
Sbjct: 442 VYFRDLWRASGTVVGIAVLVFAVFQ 516
>BI971277
Length = 700
Score = 80.5 bits (197), Expect = 1e-15
Identities = 55/175 (31%), Positives = 86/175 (48%), Gaps = 5/175 (2%)
Frame = -1
Query: 204 QLVEAGVKLHVGQDYQSVLELKFAKG----ALTIPRFEVCHWTETLLRNVVAFEQCHYPF 259
+L EAG+ L + + ++ F+ G L +P V T + N++A+E C
Sbjct: 673 ELKEAGIVLKSSKTRRP-RDVSFSYGWIRSELKLPEIVVDDTTAATVLNLIAYEMCPDFE 497
Query: 260 QTY-ITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGG 318
Y I Y LD LID DV L + I++N+LG VA + N + ++V + +
Sbjct: 496 NDYGICSYVSFLDSLIDHPDDVKALRSEQILLNSLGSDEEVANLFNTISTDLVPDMVK-- 323
Query: 319 YISLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLIQATCSI 373
Y + ++ Y D S + A+ H YFS PW I +F AA+V L LT +Q +I
Sbjct: 322 YADVRNEIEKHYSDKSRTWLALGYHTYFSNPWAIIAFHAAVVGLALTFVQTWYTI 158
>BE609719
Length = 337
Score = 77.8 bits (190), Expect = 7e-15
Identities = 48/106 (45%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Frame = -2
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
ME KL Y +F L+R+ IG I++ E +R CYS +E + + VKII D F
Sbjct: 312 MEKHKLFYSMAF-LQRSQTTSDSFIGKIEEMEPELRRCYSHTLEFSKEQLVKIIFVDCAF 136
Query: 61 IIEYFLR--SLEWPQEDPLLSKPWLRCDVKLDLILLENQLPWFVLE 104
I+E F R S EW +ED LSKP ++ DL+LLENQ+P+FVLE
Sbjct: 135 ILELFCRFGSGEW-KEDMYLSKPLTMRSMRYDLLLLENQVPFFVLE 1
>TC221646
Length = 790
Score = 71.2 bits (173), Expect = 7e-13
Identities = 53/197 (26%), Positives = 84/197 (41%), Gaps = 30/197 (15%)
Frame = +1
Query: 202 ASQLVEAGVKLHVGQDYQSVL-----ELKFAKGALTIPRFEVCHWTETLLRNVVAFEQCH 256
A++L AGV + + + ++ +++F+KG L IP V TE LRN +A+EQ
Sbjct: 94 ATRLRAAGVTIRKVERHSKLVNWFGFDIRFSKGVLEIPPLHVVDTTEVYLRNFIAWEQSR 273
Query: 257 YPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNA-VAKMINNLCLNVVQENI 315
T Y + L L+ + QD++ LVE G+++ SN + + + V + +
Sbjct: 274 IGINRQFTSYALFLRGLMCSVQDIELLVENGVLVKGTKISNRDLLTLFGTITKGV--DQM 447
Query: 316 NGGYISLCRKLN------------------------CFYEDPSHKYKAIFIHDYFSTPWK 351
+ Y LC LN C H YK I I D+ WK
Sbjct: 448 DNSYSKLCEDLNAYSAVNPLRKFPILMLHCCNLCVECIRYSCKHSYK-ILIRDHIPNVWK 624
Query: 352 ITSFAAAIVLLLLTLIQ 368
+ A LL LT++Q
Sbjct: 625 LIGIVVAAALLALTIMQ 675
>TC231575
Length = 527
Score = 70.5 bits (171), Expect = 1e-12
Identities = 40/109 (36%), Positives = 60/109 (54%), Gaps = 1/109 (0%)
Frame = +2
Query: 260 QTYITDYTILLDFLIDTSQDVDKLVEKGIMINTLGDSNAVAKMINNLCLNVVQENINGGY 319
Q +I +Y L+D LI T +DV+ LV+K I+++ LG N +A MIN LC +VV +N Y
Sbjct: 8 QPFICNYVSLIDSLIHTQEDVELLVDKEIIVHELGCHNELATMINGLCKHVV---VNCNY 178
Query: 320 I-SLCRKLNCFYEDPSHKYKAIFIHDYFSTPWKITSFAAAIVLLLLTLI 367
RKLN Y Y + YF PW+++S +V+ L ++
Sbjct: 179 YGKTSRKLNDHYNCCWKHYMGMLRSVYFRDPWRLSSTVVGVVIFLFAIV 325
>CA819940
Length = 409
Score = 65.5 bits (158), Expect = 4e-11
Identities = 48/135 (35%), Positives = 73/135 (53%)
Frame = +1
Query: 77 LLSKPWLRCDVKLDLILLENQLPWFVLEDLFNLTEPSCIDGEVSSFFDVAFHYFKVHFLQ 136
+L KPWL +V+LDL+LLENQLP+FVL+ LF L+ + S+ + K +
Sbjct: 22 ILLKPWLTTNVRLDLLLLENQLPFFVLDHLFGLS--------MQSYTSTSGRGGKKNIPP 177
Query: 137 SILPNETNNKNFTINYFHEHYQQYIMKPDQVSMQLHNLTDLLRVFYLPPDMLPKREKQIV 196
I FT +YF + + + + + + + TDLLR F+L + EK +V
Sbjct: 178 FIA--------FTFDYFSFYNRSEL---NFHGVMIKHFTDLLRTFHLQHPQQNRIEKTVV 324
Query: 197 KHLFSASQLVEAGVK 211
HL SA++L EAGV+
Sbjct: 325 -HLPSAAELSEAGVR 366
>TC230929 weakly similar to UP|Q6ZCP9 (Q6ZCP9) Pentatricopeptide (PPR)
repeat-containing protein-like, partial (11%)
Length = 831
Score = 60.5 bits (145), Expect = 1e-09
Identities = 51/208 (24%), Positives = 87/208 (41%), Gaps = 8/208 (3%)
Frame = +1
Query: 173 NLTDLLRVFYLPPDMLPKREKQIVKHLFSASQLVEAGVKLHVGQDYQSVLELKFAKGALT 232
N++ L P + + E +V ++ +L GV + ++ KG
Sbjct: 49 NMSSHLESLSSHPKLSEEEEAPVVVNIPCVRELHSVGVYFQPVEGGNMAIDFDEKKGIFY 228
Query: 233 IPRFEVCHWTETLLRNVVAFEQCHYPFQTYITDYTILLDFLIDTSQDVDKLVEKGIMINT 292
+P ++ +E ++RN+VA E P T YT L+ +IDT +DV L GI+ ++
Sbjct: 229 LPVLKLDVNSEVIMRNLVAHEALTKPDFLIFTRYTELMRGIIDTVEDVKLLKNAGIIDSS 408
Query: 293 LGDS--------NAVAKMINNLCLNVVQENINGGYISLCRKLNCFYEDPSHKYKAIFIHD 344
S N ++K I + E I +K+N +Y D + +
Sbjct: 409 SSLSVEETEELFNGMSKSIGPTKTEKLDETI--------KKVNKYYHDKRKANLYRTLTE 564
Query: 345 YFSTPWKITSFAAAIVLLLLTLIQATCS 372
Y WK+ + A VLL +T +Q CS
Sbjct: 565 YVYNSWKLFTLLATFVLLAMTALQTFCS 648
>BE346622
Length = 489
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/72 (41%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Frame = +1
Query: 1 MEDLKLRYLKSFFLERTHKGLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDACF 60
ME KL Y K+F L+RT + YI+ E R CYSE +E + VKII D+
Sbjct: 277 MEKHKLMYCKAF-LKRTKTSSDSWMSYIEGVEPKFRRCYSETLEFRKKELVKIIFVDSGX 453
Query: 61 IIEYFLRSL-EW 71
I+E RS EW
Sbjct: 454 ILELXWRSCSEW 489
>BE800256
Length = 403
Score = 42.7 bits (99), Expect = 3e-04
Identities = 34/108 (31%), Positives = 50/108 (45%), Gaps = 20/108 (18%)
Frame = +2
Query: 165 DQVSMQLHNLTDLLRVFYLPPDMLPKREKQIVKH-LFSASQLVEAGVK---------LHV 214
D+ S + TDL+R FYLP D L R H L +A++L ++GV L +
Sbjct: 86 DRYSKSTKHFTDLIRYFYLPSDWL--RSSGCALHVLRTATKLQDSGVSFEKDVERRLLDI 259
Query: 215 GQDYQSVLE----------LKFAKGALTIPRFEVCHWTETLLRNVVAF 252
D + +L L K IP+ +V H TE + RN++AF
Sbjct: 260 SFDKKPILSSFLCFGCLPYLNHFKARFRIPQLKVDHTTECVFRNLIAF 403
>AW472212
Length = 429
Score = 42.0 bits (97), Expect = 5e-04
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 3/69 (4%)
Frame = +2
Query: 2 EDLKLRYLKSFFLERTHK---GLGDCIGYIKKSEEIIRSCYSEAIEQTSDDFVKIILTDA 58
E K RY +F+ +HK L +++++ E I +CYS+ + FV +IL D+
Sbjct: 188 EKQKQRYFHAFWKRLSHKQGLALSQYKAFLEENIEKIGNCYSKPELHKEEKFVDLILLDS 367
Query: 59 CFIIEYFLR 67
FI+E FLR
Sbjct: 368 VFIMELFLR 394
>TC233543
Length = 519
Score = 39.3 bits (90), Expect = 0.003
Identities = 29/81 (35%), Positives = 43/81 (52%), Gaps = 4/81 (4%)
Frame = +2
Query: 300 AKMINNLCLNVV-QENINGGYISLCRKLNCFYEDPSHK--YKAIFIHDYFSTPWKITSFA 356
A MIN L N+ NI+ Y + KLN F+++P Y A DY T+ +
Sbjct: 2 AMMINGLMKNITTSNNISSQYFDVSEKLNAFHKNPLS*A*YLAS*ERDYX*YXLC*TAAS 181
Query: 357 -AAIVLLLLTLIQATCSIISL 376
AAI+LL+L+ +Q CSI+ +
Sbjct: 182 IAAIILLILSFVQTVCSILQV 244
>TC207591 weakly similar to UP|Q9LHW8 (Q9LHW8) ESTs AU075812(E60850), partial
(57%)
Length = 1290
Score = 29.6 bits (65), Expect = 2.3
Identities = 11/32 (34%), Positives = 20/32 (62%)
Frame = -2
Query: 329 FYEDPSHKYKAIFIHDYFSTPWKITSFAAAIV 360
F PS + +F+H YF++P + +FA++ V
Sbjct: 224 FLNQPS*MFNELFVHRYFASPASMKNFASSAV 129
>BU091636 similar to GP|6730969|dbj beta-amyrin synthase {Glycyrrhiza
glabra}, partial (18%)
Length = 436
Score = 29.3 bits (64), Expect = 3.0
Identities = 12/43 (27%), Positives = 22/43 (50%)
Frame = +3
Query: 290 INTLGDSNAVAKMINNLCLNVVQENINGGYISLCRKLNCFYED 332
+N G S + +N +C+ ++ E NGG+ + C K + D
Sbjct: 147 LNIDGHSTMFSTTLNYICMRILGEGPNGGHNNACAKARKWIHD 275
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,718,102
Number of Sequences: 63676
Number of extensions: 280937
Number of successful extensions: 1948
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1931
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1941
length of query: 377
length of database: 12,639,632
effective HSP length: 99
effective length of query: 278
effective length of database: 6,335,708
effective search space: 1761326824
effective search space used: 1761326824
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146743.5


