

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
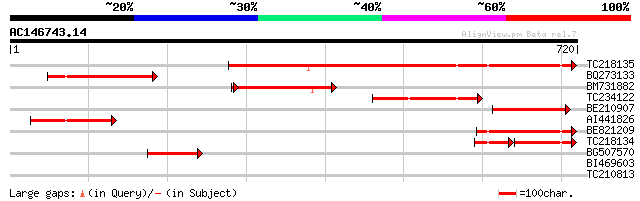
Query= AC146743.14 + phase: 0
(720 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218135 similar to UP|Q9FIL4 (Q9FIL4) Neutral ceramidase, parti... 531 e-151
BQ273133 similar to GP|12313690|db P0501G01.24 {Oryza sativa (ja... 197 2e-50
BM731882 similar to PIR|G86208|G86 protein F22G5.28 [imported] -... 183 3e-46
TC234122 similar to UP|Q93ZI6 (Q93ZI6) AT5g58980/k19m22_180, par... 171 8e-43
BE210907 171 1e-42
AI441826 similar to GP|12313690|db P0501G01.24 {Oryza sativa (ja... 168 7e-42
BE821209 111 1e-24
TC218134 weakly similar to UP|Q7XJQ9 (Q7XJQ9) At2g38010 protein,... 77 8e-24
BG507570 similar to PIR|G86208|G86 protein F22G5.28 [imported] -... 94 2e-19
BI469603 similar to GP|15982799|gb| AT5g58980/k19m22_180 {Arabid... 39 0.011
TC210813 30 3.8
>TC218135 similar to UP|Q9FIL4 (Q9FIL4) Neutral ceramidase, partial (60%)
Length = 1734
Score = 531 bits (1368), Expect = e-151
Identities = 268/447 (59%), Positives = 329/447 (72%), Gaps = 5/447 (1%)
Frame = +2
Query: 278 DGSLFVGAFCQSNVGDVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEILS 337
D FV AFCQ+N GDVSPNVLGAFCID+ PCDFNHS+C G + LC GRGPGYP+E S
Sbjct: 38 DKPRFVSAFCQTNCGDVSPNVLGAFCIDTELPCDFNHSTCGGKNELCYGRGPGYPDEFES 217
Query: 338 TKIIGERQFRSAVELFGSASEELTGKIDYRHVYLNFTNIEV---ELDNKKVVKTCPAALG 394
T+IIGERQF+ AVELF ASE++ GK+D+RH +++F+ +EV ++ +V+KTCPAA+G
Sbjct: 218 TRIIGERQFKKAVELFNGASEQIKGKVDFRHAFIDFSQLEVNPSKVGASEVIKTCPAAMG 397
Query: 395 PGFAAGTTDGPGVFGFQQGDPEISPFWKNVRDFLKEPSQYQVDCQNPKPVLLSSGEMFDP 454
FAAGTTDGPG F F+QGD + +PFWK VR+ LK P + Q+DC +PKP+LL +GEM P
Sbjct: 398 FAFAAGTTDGPGAFDFKQGDDQGNPFWKLVRNLLKTPGKEQIDCHHPKPILLDTGEMKLP 577
Query: 455 YPWAPAILPIQILRLGKLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIA 514
Y WAP+ILPIQILR+G+L+ILSVPGEFTTMAGRRLR+AVK T++S S G F + HVVIA
Sbjct: 578 YDWAPSILPIQILRVGQLVILSVPGEFTTMAGRRLRDAVK-TVLSGSKG-FGSNIHVVIA 751
Query: 515 GLTNTYSQYIATFEEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSP 574
GLTNTYSQY+ T+EEY QRYE ASTLYGPHTLSAYIQEF KLA A+ G + G P
Sbjct: 752 GLTNTYSQYVTTYEEYQVQRYEGASTLYGPHTLSAYIQEFTKLAHALISGQPV-EPGPQP 928
Query: 575 PDLLSVQKSFLLDPFGDTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLL 634
PDLL Q S L D TP G+K GD D+ P + F + D S TFWSA PR DL+
Sbjct: 929 PDLLDKQISLLTPVVMDATPIGVKFGDCSSDV--PKNSNFKRADMVSVTFWSACPRNDLM 1102
Query: 635 TEGTYAVVERLQG-ERWISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYR 692
TEGT+++VE LQG + W+ DDDD L F+W + S H A IEW IP D GVYR
Sbjct: 1103TEGTFSLVEFLQGKDMWVPAYDDDDFCLRFKWSRPFKLSSHSKATIEWRIPKDVTPGVYR 1282
Query: 693 LKHFGASKKTIVSPINYFTGASSAFAV 719
+KHFGA+ K ++ I++FTG+SSAF V
Sbjct: 1283IKHFGAA-KGLLGSIHHFTGSSSAFVV 1360
>BQ273133 similar to GP|12313690|db P0501G01.24 {Oryza sativa (japonica
cultivar-group)}, partial (16%)
Length = 420
Score = 197 bits (500), Expect = 2e-50
Identities = 100/139 (71%), Positives = 115/139 (81%)
Frame = -3
Query: 49 NIEQNTAGIHFRLRARTFIVAENLQGPRFVFVNLDAGMASQLLTIKLLERLKSRFGNLYT 108
N Q +GIHFRLRAR FIVAE G VFVNLDA MASQ++ IK++ERLK+R+G+LYT
Sbjct: 418 NTGQIASGIHFRLRARAFIVAEP-NGNWVVFVNLDACMASQIVKIKVIERLKARYGDLYT 242
Query: 109 EENVAISGIHTHAGPGGYLQYVVYSVTSLGFVTQSFDAIANAVEQSIIQAHNNLKPGSIF 168
EENVAISGIHTHAGPGGYLQYVVY VTSLGFV QSFD I N +E+ IIQAH NL+PGSIF
Sbjct: 241 EENVAISGIHTHAGPGGYLQYVVYIVTSLGFVHQSFDVIVNGIEKCIIQAHENLRPGSIF 62
Query: 169 INTGDVKEASINRSPSAYL 187
IN G++ + +NRSPSAYL
Sbjct: 61 INKGELLDGGVNRSPSAYL 5
>BM731882 similar to PIR|G86208|G86 protein F22G5.28 [imported] - Arabidopsis
thaliana, partial (16%)
Length = 422
Score = 183 bits (464), Expect(2) = 3e-46
Identities = 84/132 (63%), Positives = 103/132 (77%), Gaps = 3/132 (2%)
Frame = +3
Query: 286 FCQSNVGDVSPNVLGAFCIDSGKPCDFNHSSCNGNDLLCVGRGPGYPNEILSTKIIGERQ 345
F V DVSPNVLGAFCID+G PCDFNHS+C G + LC RGPGYP+E ST+IIGERQ
Sbjct: 15 FVNQTVEDVSPNVLGAFCIDTGLPCDFNHSTCGGKNELCYSRGPGYPDEFESTRIIGERQ 194
Query: 346 FRSAVELFGSASEELTGKIDYRHVYLNFTNIEVELDNK---KVVKTCPAALGPGFAAGTT 402
FR AV+LF +A EE+ G +D+RH Y++F+ +EV + ++ +VVKTCPAA+G FAAGTT
Sbjct: 195 FRKAVDLFNAADEEIEGDVDFRHAYIDFSQLEVTISDQGYSEVVKTCPAAMGFAFAAGTT 374
Query: 403 DGPGVFGFQQGD 414
DGPG F FQQGD
Sbjct: 375 DGPGAFDFQQGD 410
Score = 21.6 bits (44), Expect(2) = 3e-46
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = +2
Query: 282 FVGAFCQSN 290
FV AFCQSN
Sbjct: 2 FVSAFCQSN 28
>TC234122 similar to UP|Q93ZI6 (Q93ZI6) AT5g58980/k19m22_180, partial (37%)
Length = 420
Score = 171 bits (434), Expect = 8e-43
Identities = 92/140 (65%), Positives = 104/140 (73%)
Frame = +3
Query: 461 ILPIQILRLGKLIILSVPGEFTTMAGRRLREAVKETLISNSNGEFNNETHVVIAGLTNTY 520
ILPIQILR+G+LIILSVPGEFTTM GRRLR+AVK L S EF+ + H+VIAGLTNTY
Sbjct: 6 ILPIQILRIGQLIILSVPGEFTTMTGRRLRDAVKTVLTSEEYFEFD-DIHIVIAGLTNTY 182
Query: 521 SQYIATFEEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYGNGTSPPDLLSV 580
SQYI T+EEY QRYE ASTLYGPHTLSAYIQEF KLA+A+ G+ + G PPDLL
Sbjct: 183 SQYITTYEEYQVQRYEGASTLYGPHTLSAYIQEFKKLAEALVYGEPV-EPGPQPPDLLEK 359
Query: 581 QKSFLLDPFGDTTPDGIKLG 600
Q S L D TP G+ G
Sbjct: 360 QISLLPPVVVDATPLGVNFG 419
>BE210907
Length = 391
Score = 171 bits (433), Expect = 1e-42
Identities = 78/99 (78%), Positives = 87/99 (87%)
Frame = -1
Query: 614 FTKGDKPSATFWSANPRYDLLTEGTYAVVERLQGERWISVQDDDDLSLFFRWKVDNTSFH 673
F KG ATFWS+NPRYDLLTEG++AVVERLQ ERWISV DDDDLSLFFRWKVDN+S H
Sbjct: 385 FYKGRYTKATFWSSNPRYDLLTEGSFAVVERLQEERWISVYDDDDLSLFFRWKVDNSSLH 206
Query: 674 GFAAIEWEIPTDAISGVYRLKHFGASKKTIVSPINYFTG 712
G A IEWEIP DA+SG+YRLKHFGA++ TI+SPINYFTG
Sbjct: 205 GLATIEWEIPNDAVSGLYRLKHFGATRTTIISPINYFTG 89
>AI441826 similar to GP|12313690|db P0501G01.24 {Oryza sativa (japonica
cultivar-group)}, partial (15%)
Length = 412
Score = 168 bits (426), Expect = 7e-42
Identities = 84/109 (77%), Positives = 97/109 (88%)
Frame = +3
Query: 27 LIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRARTFIVAENLQGPRFVFVNLDAGM 86
LIG+GSYD+TGPAADVNMMGYAN EQ +G+HFRLRAR FIVA+ +G R VFVNLDA M
Sbjct: 87 LIGLGSYDITGPAADVNMMGYANTEQIASGVHFRLRARAFIVAQP-KGNRVVFVNLDACM 263
Query: 87 ASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGGYLQYVVYSVT 135
ASQL+ IK++ERLK+R+G+LYTE+NVAISGIHTHAGP GYLQYVVY VT
Sbjct: 264 ASQLVVIKVIERLKARYGDLYTEKNVAISGIHTHAGPAGYLQYVVYIVT 410
>BE821209
Length = 614
Score = 111 bits (277), Expect = 1e-24
Identities = 59/129 (45%), Positives = 80/129 (61%), Gaps = 2/129 (1%)
Frame = -1
Query: 593 TPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLLTEGTYAVVERLQ-GERWI 651
TP G+ GD+ D+ P + F GD +A+FWSA PR DL+TEGT+A+VE LQ + W
Sbjct: 563 TPXGVNFGDVCTDV--PRNSTFKSGDLVTASFWSACPRNDLMTEGTFALVEFLQEKDAWT 390
Query: 652 SVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKHFGASKKTIVSPINYF 710
DDDD L ++W + S A +EW IP GVYRL+HFGA+ K + I++F
Sbjct: 389 PAYDDDDFCLRYKWSRPSKLSSRSKATLEWRIPQSVAPGVYRLRHFGAA-KGLFGSIHHF 213
Query: 711 TGASSAFAV 719
TG+S+AF V
Sbjct: 212 TGSSTAFVV 186
>TC218134 weakly similar to UP|Q7XJQ9 (Q7XJQ9) At2g38010 protein, partial
(17%)
Length = 508
Score = 77.4 bits (189), Expect(2) = 8e-24
Identities = 41/81 (50%), Positives = 53/81 (64%), Gaps = 2/81 (2%)
Frame = +1
Query: 641 VVERLQG-ERWISVQDDDDLSLFFRW-KVDNTSFHGFAAIEWEIPTDAISGVYRLKHFGA 698
+VE LQG + W+ DDDD L F+W + S H A IEW IP D GVYR+KHFGA
Sbjct: 157 LVEFLQGKDTWVPAYDDDDFCLRFKWSRPFKLSSHSKATIEWRIPQDVTPGVYRIKHFGA 336
Query: 699 SKKTIVSPINYFTGASSAFAV 719
+ K ++ I++FTG+SSAF V
Sbjct: 337 A-KGLLGSIHHFTGSSSAFVV 396
Score = 52.0 bits (123), Expect(2) = 8e-24
Identities = 25/49 (51%), Positives = 31/49 (63%)
Frame = +3
Query: 591 DTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLLTEGTY 639
D TP G+K G D+ P + F +GD S TFWSA PR DL+TEGT+
Sbjct: 12 DATPIGVKXGXXXXDV--PKNSNFKRGDMVSVTFWSACPRNDLMTEGTF 152
>BG507570 similar to PIR|G86208|G86 protein F22G5.28 [imported] - Arabidopsis
thaliana, partial (15%)
Length = 426
Score = 94.4 bits (233), Expect = 2e-19
Identities = 47/69 (68%), Positives = 54/69 (78%)
Frame = +2
Query: 176 EASINRSPSAYLLNPAEERSRYPSNVDTQMTLLKFVDSASGKSKGSFSWFATHGTSMSNN 235
+A +NRSPSAYL NPA ERS++ +VD +MTLLKFVD G GSF+WFATHGTSMS
Sbjct: 5 DAGVNRSPSAYLNNPAAERSKFKYDVDKEMTLLKFVDDEWG-PLGSFNWFATHGTSMSRT 181
Query: 236 NKLISGDNK 244
N LISGDNK
Sbjct: 182 NSLISGDNK 208
>BI469603 similar to GP|15982799|gb| AT5g58980/k19m22_180 {Arabidopsis
thaliana}, partial (10%)
Length = 441
Score = 38.5 bits (88), Expect = 0.011
Identities = 16/21 (76%), Positives = 21/21 (99%)
Frame = +2
Query: 459 PAILPIQILRLGKLIILSVPG 479
P+ILPIQ+LR+G+L+ILSVPG
Sbjct: 269 PSILPIQVLRVGQLVILSVPG 331
>TC210813
Length = 914
Score = 30.0 bits (66), Expect = 3.8
Identities = 16/49 (32%), Positives = 24/49 (48%)
Frame = -1
Query: 429 KEPSQYQVDCQNPKPVLLSSGEMFDPYPWAPAILPIQILRLGKLIILSV 477
K S+ C NP P+ SSG+++ P P + LR G L L++
Sbjct: 227 KRTSRKICSCGNPSPLKFSSGQLYAPLP------KMSTLRFGNLSSLAI 99
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,556,184
Number of Sequences: 63676
Number of extensions: 391982
Number of successful extensions: 1837
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1812
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1819
length of query: 720
length of database: 12,639,632
effective HSP length: 104
effective length of query: 616
effective length of database: 6,017,328
effective search space: 3706674048
effective search space used: 3706674048
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146743.14


