

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.15 + phase: 0
(345 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
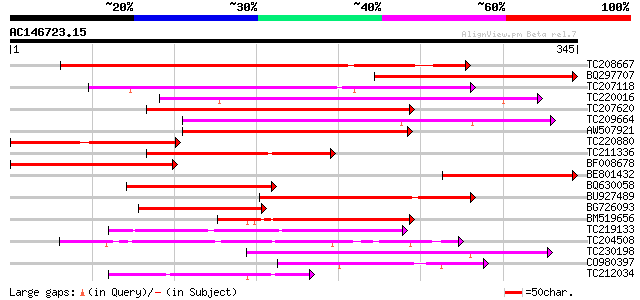
Score E
Sequences producing significant alignments: (bits) Value
TC208667 weakly similar to UP|Q84V88 (Q84V88) Cyclin D, partial ... 190 7e-49
BQ297707 180 1e-45
TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete 152 2e-37
TC220016 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (51%) 147 7e-36
TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, part... 140 6e-34
TC209664 UP|Q6T2Z7 (Q6T2Z7) Cyclin d2, partial (69%) 137 7e-33
AW507921 131 4e-31
TC220880 similar to UP|Q8GVE0 (Q8GVE0) Cyclin D1, partial (17%) 125 3e-29
TC211336 weakly similar to UP|Q8GVD9 (Q8GVD9) Cyclin D3, partial... 115 4e-26
BF008678 homologue to GP|28971972|dbj OSJNBb0041B22.15 {Oryza sa... 114 5e-26
BE801432 weakly similar to GP|6448480|emb| cyclin D1 {Antirrhinu... 106 1e-23
BQ630058 weakly similar to GP|3608179|dbj| cyclin D {Pisum sativ... 104 7e-23
BU927489 weakly similar to GP|22091622|em cyclin D2 {Daucus caro... 89 3e-18
BG726093 similar to GP|4160298|emb cyclin D2.1 protein {Nicotian... 89 3e-18
BM519656 81 6e-16
TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete 77 9e-15
TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%) 75 6e-14
TC230198 weakly similar to UP|Q7XAB7 (Q7XAB7) Cyclin D3-2, parti... 74 1e-13
CO980397 72 3e-13
TC212034 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (9%) 70 1e-12
>TC208667 weakly similar to UP|Q84V88 (Q84V88) Cyclin D, partial (24%)
Length = 1149
Score = 190 bits (483), Expect = 7e-49
Identities = 111/251 (44%), Positives = 159/251 (63%), Gaps = 2/251 (0%)
Frame = +3
Query: 32 LDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWI 91
+ SSS+ L F +E +IA ++ E +P DY R + RS++ + R +A+ WI
Sbjct: 111 MPSSSTMPLSPDPPFLCADEAAIAGLLDAEPHHMPEKDYPRRCRDRSVDVTARLDAVNWI 290
Query: 92 LKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPE-SNGWPLQLLSVACLSLAAKMEEPLVPS 150
LKVH YY F P+TA+LSVNY DRFL LP+ S GW QLLSVACLSLAAKMEE VP
Sbjct: 291 LKVHAYYEFSPVTAFLSVNYFDRFLSRCSLPQQSGGWAFQLLSVACLSLAAKMEESHVPF 470
Query: 151 LLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFII 210
LLD Q+ K++FQP+T RMEL V++ L WRLRS+TP +L +F KL S+ + + I
Sbjct: 471 LLDLQLFQPKFVFQPKTDQRMELWVMSNLKWRLRSVTPFDYLHYFFTKLPSSSSQS---I 641
Query: 211 SRATEIILSNIQDASFLTYRPSCIAAAAILSAAN-EIPNWSFVNPEHAESWCEGLSKEKI 269
+ A+ +ILS + +FL + PS +AAAA+L +AN ++P S+ + ++ E +
Sbjct: 642 TTASNLILSTTRPINFLGFAPSTVAAAAVLCSANGQLP----------LSFHDRVNDEMV 791
Query: 270 IGCYELIQEIV 280
C++L++E V
Sbjct: 792 RCCHQLMEEYV 824
>BQ297707
Length = 376
Score = 180 bits (456), Expect = 1e-45
Identities = 93/125 (74%), Positives = 106/125 (84%), Gaps = 2/125 (1%)
Frame = -1
Query: 223 DASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSS 282
+ASFL YRPSCIAAAAIL+AANEIPNWS V PE AESWC+G+ KEK+IGCY+L+QE+V +
Sbjct: 376 EASFLAYRPSCIAAAAILTAANEIPNWSVVKPEQAESWCQGIRKEKVIGCYQLMQELVIN 197
Query: 283 NNQRNAP-KVLPQLRVTARTR-RWSTVSSSSSSPSSSSSPSFSLSYKKRKLNSCFWVDVD 340
NNQR P KVLPQLRVT RTR R STVSS SS+ SSSSS SFSLS K+RKLN+ WVD +
Sbjct: 196 NNQRKLPTKVLPQLRVTTRTRMRSSTVSSFSSTSSSSSSTSFSLSCKRRKLNNRLWVDDE 17
Query: 341 KGNSE 345
KGNSE
Sbjct: 16 KGNSE 2
>TC207118 homologue to UP|Q6T2Z6 (Q6T2Z6) Cyclin d3, complete
Length = 1763
Score = 152 bits (384), Expect = 2e-37
Identities = 92/253 (36%), Positives = 139/253 (54%), Gaps = 18/253 (7%)
Frame = +3
Query: 49 EEEESIAVFIEHEFKFVPGFDYVS---------------RFQSRSLESSTREEAIAWILK 93
E+EE ++F + + + +DY + S S S R EA+ WILK
Sbjct: 399 EDEELNSLFSKEKVQHEEAYDYNNLNSDDNSNDHSNNNNNVLSDSCLSQPRREAVEWILK 578
Query: 94 VHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLD 153
V+ +YGF LTA L+V Y+DRFL S W +QL++V C+SLAAK+EE VP LLD
Sbjct: 579 VNAHYGFSALTATLAVTYLDRFLLSFHFQREKPWMIQLVAVTCISLAAKVEETQVPLLLD 758
Query: 154 FQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHF---II 210
Q++ KY+F+ +TI RMELLVL+ L W++ +TPLSFL +L G TH +
Sbjct: 759 LQVQDTKYVFEAKTIQRMELLVLSTLKWKMHPVTPLSFLDHIIRRL---GLKTHLHWEFL 929
Query: 211 SRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKII 270
R ++LS + D+ F+ PS +A A +L ++I + + ++ +SKEK+
Sbjct: 930 RRCEHLLLSVLLDSRFVGCLPSVLATATMLHVIDQIKHNGGMEYKNQLLSVLKISKEKVD 1109
Query: 271 GCYELIQEIVSSN 283
CY I ++ +N
Sbjct: 1110ECYNAILQLSKAN 1148
>TC220016 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (51%)
Length = 921
Score = 147 bits (371), Expect = 7e-36
Identities = 89/243 (36%), Positives = 129/243 (52%), Gaps = 10/243 (4%)
Frame = +1
Query: 92 LKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNG--------WPLQLLSVACLSLAAKM 143
LKV+ Y F LTA L+VNY+DRFL S N W QL +VACLSLAAK
Sbjct: 1 LKVNARYSFSTLTAVLAVNYLDRFLFSFRFQNDNNDNNNNNNPWLTQLSAVACLSLAAKF 180
Query: 144 EEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTG 203
EE VP +D Q+E +KY+F+ +T+ RME+LVL+ L W++ +TPLSFL + KL G
Sbjct: 181 EETHVPLFIDLQVEESKYLFEAKTVKRMEILVLSTLGWKMNPVTPLSFLDYITRKLGLKG 360
Query: 204 TFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEG 263
+ R ++LS D+ F+ Y PS +A A ++ N + V + G
Sbjct: 361 YLCWEFLRRCETVLLSVFADSRFMGYLPSVLATATVMRVVNTVEPRLGVEYQDQLLGILG 540
Query: 264 LSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTA--RTRRWSTVSSSSSSPSSSSSPS 321
+ KEK+ CY L+ E+VS ++ L + ++ + + V S S SSS+ S
Sbjct: 541 IDKEKVEECYNLMMEVVSGYDEEGKRSKLKKRKLESIIPCSSQNGVMEGSFSCDSSSNES 720
Query: 322 FSL 324
+ L
Sbjct: 721 WEL 729
>TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, partial (51%)
Length = 1405
Score = 140 bits (354), Expect = 6e-34
Identities = 70/164 (42%), Positives = 106/164 (63%), Gaps = 1/164 (0%)
Frame = +3
Query: 84 REEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKM 143
R EA+ W+LKV+ +Y F LTA LSVNY DRFL S W +QL +VACLS+AAK+
Sbjct: 279 RREAVEWMLKVNSHYSFSALTAVLSVNYFDRFLFSFRFQNDKPWMVQLAAVACLSIAAKV 458
Query: 144 EEPLVPSLLDF-QIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDST 202
EE VP L+D Q++ ++Y+F+ +TI +ME+LVL+ L W++ TPLSFL +F +L S
Sbjct: 459 EETHVPFLIDLQQVDESRYLFEAKTIKKMEILVLSTLGWKMNPPTPLSFLDYFTRRLGSK 638
Query: 203 GTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEI 246
+S++ ++LS + D+ F++Y PS +A A ++ +
Sbjct: 639 DHLCWEFLSKSQGVLLSLLGDSRFMSYLPSVLATATMMHVVKSV 770
>TC209664 UP|Q6T2Z7 (Q6T2Z7) Cyclin d2, partial (69%)
Length = 903
Score = 137 bits (345), Expect = 7e-33
Identities = 88/239 (36%), Positives = 136/239 (56%), Gaps = 12/239 (5%)
Frame = +2
Query: 106 YLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQP 165
YLS+NY+DRFL + LP+ W +QLL+VAC+SLAAK++E VP LD Q+ +K++F+
Sbjct: 8 YLSINYLDRFLCAYELPKGRVWTMQLLAVACVSLAAKLDETEVPLSLDLQVGESKFLFEA 187
Query: 166 RTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDAS 225
+TI RMELLVL+ L WR+++ITP +FL +F CK++ + I R+ ++I S +
Sbjct: 188 KTIQRMELLVLSTLKWRMQAITPFTFLDYFLCKINDDQSPLRSSIMRSIQLISSTARGID 367
Query: 226 FLTYRPSCIAAA------AILSAANEIPNWSFVNPEHAES-WCEGLSKEKIIGCYELIQE 278
FL ++PS IAAA A A + V+ A S + + KE+++ C ++IQE
Sbjct: 368 FLEFKPSEIAAAVKPSEIAAAVAMYVMGETQTVDTGKAISVLIQHVEKERLLKCVQMIQE 547
Query: 279 IV----SSNNQRNAPKVLPQLRVTARTRRWSTVSSSSSSPSSSSSPSF-SLSYKKRKLN 332
+ S+ + + LPQ + S ++ SS + S S K+RKLN
Sbjct: 548 LSCNSGSAKDSSASVTCLPQSPIGVLDALCFNYKSDDTNASSCVNSSHNSPVAKRRKLN 724
>AW507921
Length = 445
Score = 131 bits (330), Expect = 4e-31
Identities = 65/140 (46%), Positives = 95/140 (67%)
Frame = +2
Query: 106 YLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQP 165
YLS+NY+DRFL + LP+ W +QLL+VACLSLAAK++E VP LD Q+ +K++F+
Sbjct: 5 YLSINYLDRFLFAYELPKGRVWTMQLLAVACLSLAAKLDETEVPLSLDLQVGESKFLFEA 184
Query: 166 RTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDAS 225
+TI RMELLVL+ L WR+++ITP +FL +F CK++ + I R+ ++I S +
Sbjct: 185 KTIQRMELLVLSTLKWRMQAITPFTFLDYFLCKINDDQSPLRSSIMRSIQLISSTARGID 364
Query: 226 FLTYRPSCIAAAAILSAANE 245
FL ++PS IAAA + E
Sbjct: 365 FLEFKPSEIAAAVAMYVMGE 424
>TC220880 similar to UP|Q8GVE0 (Q8GVE0) Cyclin D1, partial (17%)
Length = 454
Score = 125 bits (314), Expect = 3e-29
Identities = 64/105 (60%), Positives = 80/105 (75%), Gaps = 1/105 (0%)
Frame = +1
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECS-SDLDSSSSSQLPSSSLFAEEEEESIAVFIE 59
M +S SD +LLCGEDSS +L+G+ PECS SD+DSS P++ E+ SIA FIE
Sbjct: 154 MSVSCLSDYDLLCGEDSSGILSGESPECSFSDIDSSPPPPSPTT-----EDCYSIASFIE 318
Query: 60 HEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLT 104
HE FVPGF+Y+SRFQSRSL+++ REE++ WILKVH YYGFQPLT
Sbjct: 319 HERNFVPGFEYLSRFQSRSLDANAREESVGWILKVHAYYGFQPLT 453
>TC211336 weakly similar to UP|Q8GVD9 (Q8GVD9) Cyclin D3, partial (28%)
Length = 1351
Score = 115 bits (287), Expect = 4e-26
Identities = 60/115 (52%), Positives = 76/115 (65%)
Frame = +2
Query: 84 REEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKM 143
R EAI WILK GF+ TAYLSV Y DRFL R + W ++LLS+ACLSLAAKM
Sbjct: 65 RVEAINWILKTRATLGFRFETAYLSVTYFDRFLSRRSIDSEKSWAIRLLSIACLSLAAKM 244
Query: 144 EEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACK 198
EE VP L +F+++ Y F+ + I +MELLVL+ L+W + ITP FLS+F K
Sbjct: 245 EECNVPGLSEFKLD--DYSFEGKVIQKMELLVLSTLEWEMGIITPFDFLSYFITK 403
>BF008678 homologue to GP|28971972|dbj OSJNBb0041B22.15 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 313
Score = 114 bits (286), Expect = 5e-26
Identities = 62/103 (60%), Positives = 73/103 (70%), Gaps = 1/103 (0%)
Frame = +2
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECSS-DLDSSSSSQLPSSSLFAEEEEESIAVFIE 59
M +S SD +LLCGEDSS VL+G+ PECSS D+ SS P S E+ SIA FIE
Sbjct: 5 MSVSCLSDYDLLCGEDSSGVLSGESPECSSSDIGSSPLPPSPPPSPTMTEDCYSIASFIE 184
Query: 60 HEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQP 102
HE FVPGF+Y+SRF SRSL++ REE+ AWI KVH YYGFQP
Sbjct: 185 HERYFVPGFEYLSRFLSRSLDAYAREESGAWISKVHAYYGFQP 313
>BE801432 weakly similar to GP|6448480|emb| cyclin D1 {Antirrhinum majus},
partial (10%)
Length = 287
Score = 106 bits (265), Expect = 1e-23
Identities = 59/84 (70%), Positives = 69/84 (81%), Gaps = 2/84 (2%)
Frame = +1
Query: 264 LSKEKIIGCYELIQEIVSSNNQRNAP-KVLPQLRVTARTR-RWSTVSSSSSSPSSSSSPS 321
L +EK+IGCY+L+QE+V +NNQR P KVLPQLRVT RTR R STVSS SS+ SSSSS S
Sbjct: 34 LLQEKVIGCYQLMQELVINNNQRKLPTKVLPQLRVTTRTRMRSSTVSSFSSTSSSSSSTS 213
Query: 322 FSLSYKKRKLNSCFWVDVDKGNSE 345
FSLS K+RKLN+ WVD +KGNSE
Sbjct: 214 FSLSCKRRKLNNRLWVDDEKGNSE 285
>BQ630058 weakly similar to GP|3608179|dbj| cyclin D {Pisum sativum}, partial
(19%)
Length = 433
Score = 104 bits (259), Expect = 7e-23
Identities = 51/91 (56%), Positives = 64/91 (70%)
Frame = +2
Query: 72 SRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQL 131
+ F ++ + REEAI WILKVH YY F+P TAYLSV+Y +RFL S + WPLQL
Sbjct: 161 NNFAKKTWLINAREEAINWILKVHAYYSFKPETAYLSVDYFNRFLLSHTFTQDKAWPLQL 340
Query: 132 LSVACLSLAAKMEEPLVPSLLDFQIEGAKYI 162
LSV CLSLAAKMEE VP LLD Q+ ++++
Sbjct: 341 LSVTCLSLAAKMEESKVPLLLDLQVIESRFL 433
>BU927489 weakly similar to GP|22091622|em cyclin D2 {Daucus carota}, partial
(27%)
Length = 449
Score = 89.0 bits (219), Expect = 3e-18
Identities = 51/132 (38%), Positives = 82/132 (61%), Gaps = 1/132 (0%)
Frame = +3
Query: 153 DFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISR 212
D Q+ +KYIF+ +TI RMELLVL+ L WR+++ITP SF+ F K++ + I +
Sbjct: 3 DLQVGESKYIFEAKTIQRMELLVLSTLRWRMQAITPFSFIDHFLYKINDDQSPIGASILQ 182
Query: 213 ATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAES-WCEGLSKEKIIG 271
+ ++ILS ++ FL +RPS IAAA +S E V+ E A S + + KE+++
Sbjct: 183 SIQLILSTVRGIDFLEFRPSEIAAAVAISVVGE---GQTVHTEKAISVLIQLVEKERVLK 353
Query: 272 CYELIQEIVSSN 283
C ++IQE+ S++
Sbjct: 354 CVKMIQELASNS 389
>BG726093 similar to GP|4160298|emb cyclin D2.1 protein {Nicotiana tabacum},
partial (29%)
Length = 412
Score = 89.0 bits (219), Expect = 3e-18
Identities = 42/78 (53%), Positives = 53/78 (67%)
Frame = +3
Query: 79 LESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLS 138
L+ S R+EA+ WI K H Y+ F P + LSVNY+DRFL LP W +QLL+VACLS
Sbjct: 3 LDLSVRKEALDWIWKAHAYFDFGPCSLCLSVNYLDRFLSVYELPRGKSWSMQLLAVACLS 182
Query: 139 LAAKMEEPLVPSLLDFQI 156
+AAKMEE VP +D Q+
Sbjct: 183 IAAKMEEIKVPPCVDLQV 236
Score = 30.8 bits (68), Expect = 0.94
Identities = 14/44 (31%), Positives = 25/44 (56%)
Frame = +1
Query: 139 LAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWR 182
LA+ + + ++ + K+ F+ + I RMELLVL+ L W+
Sbjct: 280 LASTFSSDFITFVCNWLVGEPKFAFEAKDIQRMELLVLSTLRWK 411
>BM519656
Length = 428
Score = 81.3 bits (199), Expect = 6e-16
Identities = 52/126 (41%), Positives = 80/126 (63%), Gaps = 6/126 (4%)
Frame = +2
Query: 127 WPLQLLSVACLSLAAKM---EEPL--VPSLLDFQIEGAKYIFQPRTILRMELLVLTILDW 181
W L+L++V+C+SLA KM E P V +LL+ Q +G IF+ +TI RME L+L L W
Sbjct: 14 WVLRLIAVSCISLAVKMMRTEYPFTDVQALLN-QSDGG-IIFETQTIQRMEALILGALQW 187
Query: 182 RLRSITPLSFLSFFACKLDSTG-TFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
R+RSITP SF++FF + + +RA+EII + ++ ++PS IAA+A+L
Sbjct: 188 RMRSITPFSFVAFFIALMGLKDLPMGQVLKNRASEIIFKSQREIRLWGFKPSIIAASALL 367
Query: 241 SAANEI 246
A++E+
Sbjct: 368 CASHEL 385
>TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete
Length = 1804
Score = 77.4 bits (189), Expect = 9e-15
Identities = 59/183 (32%), Positives = 88/183 (47%), Gaps = 1/183 (0%)
Frame = +2
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E K P D++ + Q + + SS R I W+++V E Y P T YL+VNY+DR+L
Sbjct: 734 EAKKRPSTDFMEKIQ-KEINSSMRAILIDWLVEVAEEYRLVPDTLYLTVNYIDRYLSGNV 910
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
+ N LQLL VA + +A+K EE P + +F F+ +L+ME VL L
Sbjct: 911 M---NRQRLQLLGVASMMIASKYEEICAPQVEEFCYITDNTYFK-EEVLQMESAVLNFLK 1078
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILS-NIQDASFLTYRPSCIAAAAI 239
+ + + T FL F + T I ++ + S L Y PS +AA+AI
Sbjct: 1079FEMTAPTVKCFLRRFVRAAQGVDEVPSLQLECLTNYIAELSLMEYSMLGYAPSLVAASAI 1258
Query: 240 LSA 242
A
Sbjct: 1259FLA 1267
>TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%)
Length = 1283
Score = 74.7 bits (182), Expect = 6e-14
Identities = 71/256 (27%), Positives = 118/256 (45%), Gaps = 10/256 (3%)
Frame = +2
Query: 31 DLDSSS-SSQLPSSSLFAEEEEESIAVF---IEHEFKFVPGFDYVSRFQSRSLESSTREE 86
D+D+S ++L + L A E + I F +E+E + DY+ + R
Sbjct: 263 DIDASDVDNELAAVELAAVEYIDDIYKFYKLVENESR---PHDYIG--SQPEINERMRAI 427
Query: 87 AIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEP 146
+ W++ VH + T YL++N +DRFL + +P LQL+ ++ + +A+K EE
Sbjct: 428 LVDWLIDVHTKFELSLETLYLTINIIDRFLAVKTVPRRE---LQLVGISAMLMASKYEEI 598
Query: 147 LVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFF---ACKLDSTG 203
P + DF + + IL ME +L L+W L TP FL F A
Sbjct: 599 WPPEVNDFVCLSDR-AYTHEQILAMEKTILNKLEWTLTVPTPFVFLVRFIKAAVPDQELE 775
Query: 204 TFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA---ANEIPNWSFVNPEHAESW 260
HF+ +E+ + N + L Y PS +AA+A+ +A N+ P W+ H
Sbjct: 776 NMAHFM----SELGMMNY---ATLMYCPSMVAASAVFAARCTLNKAPLWNETLKLHT--- 925
Query: 261 CEGLSKEKIIGCYELI 276
G S+E+++ C L+
Sbjct: 926 --GYSQEQLMDCARLL 967
>TC230198 weakly similar to UP|Q7XAB7 (Q7XAB7) Cyclin D3-2, partial (56%)
Length = 913
Score = 73.9 bits (180), Expect = 1e-13
Identities = 54/193 (27%), Positives = 93/193 (47%), Gaps = 7/193 (3%)
Frame = +1
Query: 145 EPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGT 204
E VP Q+E ++++F +TI RMELLVL+ L WR+ +TP+ +L
Sbjct: 1 ETHVPLXXXXQVEESRFVFXXKTIQRMELLVLSTLKWRMHPVTPIXXFEHIVRRLGLKSR 180
Query: 205 FTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGL 264
+ R ++L+ I D+ ++Y PS +AAA ++ EI +++ +
Sbjct: 181 LHWEFLWRCERVLLNVIADSRVMSYLPSTLAAATMIRVIKEIESFNATEYIDQLLGLLKI 360
Query: 265 SKEKIIGCYELIQEI------VSSNNQRNAPKVLPQLRVTARTRRWSTVSSSSS-SPSSS 317
S+E++ CY++IQ++ + S +Q+ P +S SS+ S + SSS
Sbjct: 361 SEEQVNQCYKIIQKLLGCYEGIYSLHQKRKRLSEPGSPGAVTDASFSCDSSNDSWTVSSS 540
Query: 318 SSPSFSLSYKKRK 330
S S K+RK
Sbjct: 541 VSLSLEPLLKRRK 579
>CO980397
Length = 664
Score = 72.4 bits (176), Expect = 3e-13
Identities = 46/134 (34%), Positives = 75/134 (55%), Gaps = 6/134 (4%)
Frame = -3
Query: 164 QPRTILRMELLVLTILDWRLRSITPLSFLSFFACKL---DSTGTFTHFIISRATEIILSN 220
+P+T+ RMELLV+ L WRLR+ITP F+ F KL ST +I+S +++I+
Sbjct: 662 KPKTVQRMELLVMASLKWRLRTITPFDFVHLFISKLLCSASTWGDLSYIVSLVSDVIIRT 483
Query: 221 IQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWC--EGLSKEKIIGCYELI-Q 277
FL + PS IAAAA+L N+ + +S+C + +S E + CY+L+ Q
Sbjct: 482 CLVMDFLEFSPSTIAAAALLWVTNQC-------VDDKKSYCLHKNISIEMVKKCYKLMKQ 324
Query: 278 EIVSSNNQRNAPKV 291
+++ ++ PK+
Sbjct: 323 KLIIRRSELYWPKI 282
>TC212034 similar to UP|Q7XAB6 (Q7XAB6) Cyclin D3-1, partial (9%)
Length = 698
Score = 70.1 bits (170), Expect = 1e-12
Identities = 45/130 (34%), Positives = 72/130 (54%), Gaps = 5/130 (3%)
Frame = +1
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E +P +Y ++ + S R + ++ I ++ F P+ YL++NY+DRFL ++
Sbjct: 319 ESDHIPPPNYCQSLKASDFDISVRRDVVSLISQLS--CTFDPVLPYLAINYLDRFLANQG 492
Query: 121 LPESNGWPLQLLSVACLSLAAKM-----EEPLVPSLLDFQIEGAKYIFQPRTILRMELLV 175
+ + W +LL+V+C SLAAKM V L++ GA IF+ +TI RME +V
Sbjct: 493 ILQPKPWANKLLAVSCFSLAAKMLKTEYSATDVQVLMNHGDGGA--IFETQTIQRMEGIV 666
Query: 176 LTILDWRLRS 185
L L WR+RS
Sbjct: 667 LGALQWRMRS 696
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,522,769
Number of Sequences: 63676
Number of extensions: 281114
Number of successful extensions: 5047
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 3183
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4234
length of query: 345
length of database: 12,639,632
effective HSP length: 98
effective length of query: 247
effective length of database: 6,399,384
effective search space: 1580647848
effective search space used: 1580647848
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146723.15


