

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146722.7 + phase: 1 /pseudo/partial
(293 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
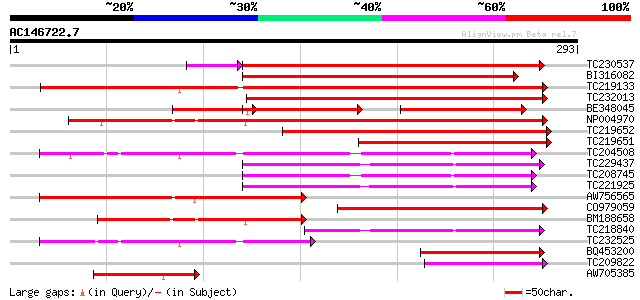
Score E
Sequences producing significant alignments: (bits) Value
TC230537 mitotic cyclin a2-type 236 5e-64
BI316082 similar to PIR|T07672|T07 cyclin a2-type mitosis-speci... 225 2e-59
TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete 215 2e-56
TC232013 similar to UP|Q9S957 (Q9S957) Cyclin A homolog, partial... 202 1e-52
BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-speci... 105 1e-52
NP004970 mitotic cyclin a1-type 191 3e-49
TC219652 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, p... 177 6e-45
TC219651 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, p... 152 1e-37
TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%) 117 5e-27
TC229437 similar to UP|Q40670 (Q40670) Cyclin (Fragment), partia... 108 2e-24
TC208745 homologue to UP|CG22_SOYBN (P25012) G2/mitotic-specific... 102 2e-22
TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 100 8e-22
AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-speci... 94 7e-20
CO979059 91 8e-19
BM188658 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 77 7e-15
TC218840 similar to UP|CG1B_MEDVA (P46277) G2/mitotic-specific c... 73 1e-13
TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type,... 64 1e-10
BQ453200 similar to GP|7242793|emb| cyclin A3.1 {Pisum sativum},... 61 7e-10
TC209822 similar to UP|Q9M4K9 (Q9M4K9) Cyclin A3.1, partial (24%) 59 3e-09
AW705385 similar to PIR|T07672|T076 cyclin a2-type mitosis-spec... 58 6e-09
>TC230537 mitotic cyclin a2-type
Length = 993
Score = 236 bits (603), Expect(2) = 5e-64
Identities = 116/156 (74%), Positives = 134/156 (85%)
Frame = +1
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEE+CAP VE+FC ITDNTY+++EVLKME+EVL+L+HFQL+VPTIKTFLRRFIQ AQ+S
Sbjct: 103 YEEMCAPRVEEFCFITDNTYTKEEVLKMEREVLDLVHFQLSVPTIKTFLRRFIQAAQSSY 282
Query: 181 KVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTN 240
K +LEFLANYLAELALVE +F QF PS +AAS+V LA+WTLN+SEHPWNPTL+HYT
Sbjct: 283 KAPCVELEFLANYLAELALVECNFFQFLPSLVAASAVFLAKWTLNESEHPWNPTLKHYTK 462
Query: 241 YKASELKTTVLELIDLQLNTKRCRLNAVREKYQHQK 276
YKASELKT VL L DLQLNTK LNAV EKY+ QK
Sbjct: 463 YKASELKTVVLALQDLQLNTKGSSLNAVPEKYKQQK 570
Score = 25.8 bits (55), Expect(2) = 5e-64
Identities = 14/29 (48%), Positives = 16/29 (54%)
Frame = +3
Query: 92 ESH*SIPFPTFDYQTKAAVAWYYLHADFI 120
E H*S+ + KA VA YLHAD I
Sbjct: 12 EPH*SVSLNKTHSEAKAPVARCYLHADCI 98
>BI316082 similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (31%)
Length = 456
Score = 225 bits (574), Expect = 2e-59
Identities = 111/143 (77%), Positives = 123/143 (85%)
Frame = +3
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEEICAP VE+FC ITDNTY++ EVLKME EVLNLLHFQL+VPT KTFLRRFI +++S
Sbjct: 24 YEEICAPRVEEFCFITDNTYTKAEVLKMESEVLNLLHFQLSVPTTKTFLRRFILASESSY 203
Query: 181 KVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTN 240
KV+ +LEFLANYLAEL LVEYSFLQF PS IAAS+VLLARWTLNQSEHPWN T+EHYTN
Sbjct: 204 KVSYVELEFLANYLAELTLVEYSFLQFLPSLIAASAVLLARWTLNQSEHPWNSTMEHYTN 383
Query: 241 YKASELKTTVLELIDLQLNTKRC 263
YK SELKTTVL L DLQ + K C
Sbjct: 384 YKVSELKTTVLALADLQHDMKGC 452
>TC219133 UP|Q39879 (Q39879) Mitotic cyclin a2-type, complete
Length = 1804
Score = 215 bits (548), Expect = 2e-56
Identities = 115/265 (43%), Positives = 166/265 (62%), Gaps = 3/265 (1%)
Frame = +2
Query: 17 VDIDSKLRDSPIWTSYAPDIYTNIHVRECERRPLANYMETLQQDITPGMRGILVDWLVEV 76
V++D+ D + ++A DIY ++ E ++RP ++ME +Q++I MR IL+DWLVEV
Sbjct: 653 VNVDNNYADPQLCATFACDIYKHLRASEAKKRPSTDFMEKIQKEINSSMRAILIDWLVEV 832
Query: 77 ADEFKLVPRH---TLPCCESH*SIPFPTFDYQTKAAVAWYYLHADFIYEEICAPGVEDFC 133
A+E++LVP T+ + + S VA + + YEEICAP VE+FC
Sbjct: 833 AEEYRLVPDTLYLTVNYIDRYLSGNVMNRQRLQLLGVASMMIASK--YEEICAPQVEEFC 1006
Query: 134 VITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSSKVAQADLEFLANY 193
ITDNTY ++EVL+ME VLN L F++ PT+K FLRRF++ AQ +V LE L NY
Sbjct: 1007YITDNTYFKEEVLQMESAVLNFLKFEMTAPTVKCFLRRFVRAAQGVDEVPSLQLECLTNY 1186
Query: 194 LAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLEL 253
+AEL+L+EYS L + PS +AAS++ LA++ L S+ PWN TL+HYT Y+ S+L V +L
Sbjct: 1187IAELSLMEYSMLGYAPSLVAASAIFLAKFILFPSKKPWNSTLQHYTLYQPSDLCVCVKDL 1366
Query: 254 IDLQLNTKRCRLNAVREKYQHQKVR 278
L N+ L A+REKY K +
Sbjct: 1367HRLCCNSPNSNLPAIREKYSQHKYK 1441
>TC232013 similar to UP|Q9S957 (Q9S957) Cyclin A homolog, partial (38%)
Length = 814
Score = 202 bits (515), Expect = 1e-52
Identities = 100/156 (64%), Positives = 120/156 (76%)
Frame = +1
Query: 123 EICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSSKV 182
EI AP +EDFC ITDNTY++ EVLKME++VL +QL PTI+TF+RRF++ AQ S K
Sbjct: 1 EINAPRIEDFCFITDNTYTKAEVLKMERQVLKSSEYQLFAPTIQTFVRRFLRAAQASYKD 180
Query: 183 AQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYK 242
+LE+LANYLAEL L++Y FL F PS IAAS+V LARWTL+QS HPWNPTL+HY YK
Sbjct: 181 QSLELEYLANYLAELTLMDYGFLNFLPSIIAASAVFLARWTLDQSNHPWNPTLQHYACYK 360
Query: 243 ASELKTTVLELIDLQLNTKRCRLNAVREKYQHQKVR 278
AS+LKTTVL L DLQLNT C L AVR KY+ K +
Sbjct: 361 ASDLKTTVLALQDLQLNTDGCPLTAVRTKYRQDKFK 468
>BE348045 similar to PIR|T07672|T07 cyclin a2-type mitosis-specific -
soybean, partial (34%)
Length = 491
Score = 105 bits (263), Expect(3) = 1e-52
Identities = 51/65 (78%), Positives = 55/65 (84%)
Frame = -2
Query: 203 SFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTKR 262
SFLQF PS IAAS+VLLARWTLNQSEHPWN T+EHYTNYK SELKTTVL L DLQ + K
Sbjct: 196 SFLQFLPSLIAASAVLLARWTLNQSEHPWNSTMEHYTNYKVSELKTTVLALADLQHDMKG 17
Query: 263 CRLNA 267
C LN+
Sbjct: 16 CSLNS 2
Score = 99.4 bits (246), Expect(3) = 1e-52
Identities = 47/62 (75%), Positives = 54/62 (86%)
Frame = -1
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEEICAP VE+FC ITDNTY++ EVLKME EVLNLLHFQL+VPT KTFLRRFI +Q+S
Sbjct: 377 YEEICAPRVEEFCFITDNTYTKAEVLKMESEVLNLLHFQLSVPTTKTFLRRFILASQSSY 198
Query: 181 KV 182
K+
Sbjct: 197 KL 192
Score = 40.0 bits (92), Expect(3) = 1e-52
Identities = 22/46 (47%), Positives = 29/46 (62%), Gaps = 2/46 (4%)
Frame = -3
Query: 85 RHTLPCCESH*SIPFPTFDYQTKAAVAWYYLHADFIY--EEICAPG 128
R++LP ES *S+ F F +TKA VA YLHAD I ++C+ G
Sbjct: 489 RYSLPYSESD*SVSFSKFGSETKAPVAGCYLHADCIKV*RDLCSSG 352
>NP004970 mitotic cyclin a1-type
Length = 1047
Score = 191 bits (486), Expect = 3e-49
Identities = 109/253 (43%), Positives = 155/253 (61%), Gaps = 5/253 (1%)
Frame = +1
Query: 31 SYAPDIYTNIHVRECE--RRPLANYMETLQQDITPGMRGILVDWLVEVADEFKLVPRHTL 88
SY DI+ + E + RRP+ NY+E Q+ +TP MRGILVDWLVEVA+E+KL+ TL
Sbjct: 235 SYVSDIHEYLREMEMQKKRRPMVNYIEKFQKIVTPTMRGILVDWLVEVAEEYKLLS-DTL 411
Query: 89 PCCESH*SIPFPTFDYQTKAAVAWYYLHADFI---YEEICAPGVEDFCVITDNTYSRQEV 145
S+ F + + TK+ + + + I YEE P V++FC ITDNTY + EV
Sbjct: 412 HLSVSYID-RFLSVNPVTKSRLQLLGVSSMLIAAKYEETDPPSVDEFCSITDNTYDKAEV 588
Query: 146 LKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSSKVAQADLEFLANYLAELALVEYSFL 205
+KME ++L L F++ PT+ TFLRR+ VA K + +E L +Y+ EL+L++Y L
Sbjct: 589 VKMEADILKSLKFEMGNPTVSTFLRRYANVASDVQKTPNSQIEHLGSYIGELSLLDYDCL 768
Query: 206 QFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTKRCRL 265
+F PS +AAS + LA++ + HPW +L + YK +ELK VL L DL L+ K
Sbjct: 769 RFLPSIVAASVIFLAKFIIWPEVHPWTSSLCECSGYKPAELKECVLILHDLYLSRKAASF 948
Query: 266 NAVREKYQHQKVR 278
AVREKY+HQK +
Sbjct: 949 KAVREKYKHQKFK 987
>TC219652 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, partial (30%)
Length = 670
Score = 177 bits (448), Expect = 6e-45
Identities = 90/139 (64%), Positives = 107/139 (76%)
Frame = +1
Query: 142 RQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSSKVAQADLEFLANYLAELALVE 201
R+ + E+E FQL+VPT KTFLRRFI +Q+S KV+ +LEFLANYLAEL LVE
Sbjct: 1 RERERERERERERERDFQLSVPTTKTFLRRFILASQSSYKVSYVELEFLANYLAELTLVE 180
Query: 202 YSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTK 261
YSFLQF PS IAAS+VLLARWTLNQSEHPWN T+EHYTNYK SELKT VL L DL + K
Sbjct: 181 YSFLQFLPSLIAASAVLLARWTLNQSEHPWNSTMEHYTNYKVSELKTAVLALADLLHDMK 360
Query: 262 RCRLNAVREKYQHQKVRNL 280
C ++++REKY QK R++
Sbjct: 361 GCSVHSIREKYTQQKFRSV 417
>TC219651 similar to UP|Q39878 (Q39878) Mitotic cyclin a2-type, partial (24%)
Length = 701
Score = 152 bits (385), Expect = 1e-37
Identities = 74/100 (74%), Positives = 85/100 (85%)
Frame = +2
Query: 181 KVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTN 240
+V+ +LEFLANYLAEL LVEYSFLQF PS IAAS+VLLARWTLNQSEHPWN T+EHYTN
Sbjct: 113 QVSYVELEFLANYLAELTLVEYSFLQFLPSLIAASAVLLARWTLNQSEHPWNSTMEHYTN 292
Query: 241 YKASELKTTVLELIDLQLNTKRCRLNAVREKYQHQKVRNL 280
YK SELKTTVL L DLQ + K C LN++REKY+ QK R++
Sbjct: 293 YKVSELKTTVLALADLQHDMKGCSLNSIREKYKQQKFRSV 412
>TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%)
Length = 1283
Score = 117 bits (294), Expect = 5e-27
Identities = 89/267 (33%), Positives = 135/267 (50%), Gaps = 10/267 (3%)
Frame = +2
Query: 16 IVDIDSKLRDSPIWT------SYAPDIYTNIHVRECERRPLANYMETLQQDITPGMRGIL 69
I+DID+ D+ + Y DIY + E E RP +Y+ + Q +I MR IL
Sbjct: 257 IIDIDASDVDNELAAVELAAVEYIDDIYKFYKLVENESRP-HDYIGS-QPEINERMRAIL 430
Query: 70 VDWLVEVADEFKLVPRH---TLPCCESH*SIP-FPTFDYQTKAAVAWYYLHADFIYEEIC 125
VDWL++V +F+L T+ + ++ P + Q A YEEI
Sbjct: 431 VDWLIDVHTKFELSLETLYLTINIIDRFLAVKTVPRRELQLVGISAMLMASK---YEEIW 601
Query: 126 APGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSSKVAQA 185
P V DF ++D Y+ +++L MEK +LN L + L VPT FL RFI+ A V
Sbjct: 602 PPEVNDFVCLSDRAYTHEQILAMEKTILNKLEWTLTVPTPFVFLVRFIKAA-----VPDQ 766
Query: 186 DLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTNYKASE 245
+LE +A++++EL ++ Y+ L + PS +AAS+V AR TLN++ WN TL+ +T Y +
Sbjct: 767 ELENMAHFMSELGMMNYATLMYCPSMVAASAVFAARCTLNKAP-LWNETLKLHTGYSQEQ 943
Query: 246 LKTTVLELIDLQLNTKRCRLNAVREKY 272
L L+ +L V KY
Sbjct: 944 LMDCARLLVGFHSTLGNGKLRVVYRKY 1024
>TC229437 similar to UP|Q40670 (Q40670) Cyclin (Fragment), partial (77%)
Length = 830
Score = 108 bits (271), Expect = 2e-24
Identities = 62/156 (39%), Positives = 89/156 (56%)
Frame = +1
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEE+ P VEDF +ITD Y+R EVL MEK ++N+L F+L+VPT F+RRF++ A +
Sbjct: 70 YEEVSVPTVEDFILITDKAYTRNEVLDMEKLMMNILQFKLSVPTPYMFMRRFLKAAHSDK 249
Query: 181 KVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTN 240
K LE L+ +L EL LVE L+F PS +AA+++ A+ +L Q + W T E YT+
Sbjct: 250 K-----LELLSFFLVELCLVECKMLKFSPSLLAAAAIYTAQCSLYQFKQ-WTKTTEWYTD 411
Query: 241 YKASELKTTVLELIDLQLNTKRCRLNAVREKYQHQK 276
Y +L ++ +L V KY K
Sbjct: 412 YSEEKLLECSRLMVTFHQKAGSGKLTGVYRKYNTWK 519
>TC208745 homologue to UP|CG22_SOYBN (P25012) G2/mitotic-specific cyclin
S13-7 (B-like cyclin) (Fragment), partial (74%)
Length = 879
Score = 102 bits (255), Expect = 2e-22
Identities = 58/152 (38%), Positives = 87/152 (57%)
Frame = +1
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEEI P V DF ++D Y+ +++L MEK +LN L + L VPT FL RFI+ A
Sbjct: 58 YEEIWPPEVNDFVCLSDRAYTHEQILAMEKTILNKLEWTLTVPTPFVFLVRFIKAA---- 225
Query: 181 KVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYTN 240
V +LE +A++++EL ++ Y+ L + PS +AAS+V AR TLN++ WN TL+ +T
Sbjct: 226 -VPDQELENMAHFMSELGMMNYATLMYCPSMVAASAVFAARCTLNKAP-LWNETLKLHTG 399
Query: 241 YKASELKTTVLELIDLQLNTKRCRLNAVREKY 272
Y +L L+ + +L V KY
Sbjct: 400 YSQEQLMDCARLLVGFHSTLENGKLRVVYRKY 495
>TC221925 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(46%)
Length = 802
Score = 100 bits (249), Expect = 8e-22
Identities = 61/153 (39%), Positives = 89/153 (57%), Gaps = 1/153 (0%)
Frame = +2
Query: 121 YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVLNLLHFQLAVPTIKTFLRRFIQVAQTSS 180
YEEI AP V DF I+DN Y Q+VL ME +L L + L VPT FL R+I+ + S
Sbjct: 116 YEEIWAPEVNDFECISDNAYVSQQVLMMEXXILRKLEWYLTVPTPYHFLVRYIKASTPSD 295
Query: 181 KVAQADLEFLANYLAELALVEY-SFLQFQPSKIAASSVLLARWTLNQSEHPWNPTLEHYT 239
K ++E + +LAEL L+ Y + + ++PS IAA++V AR TL +S W TL+HYT
Sbjct: 296 K----EMENMVFFLAELGLMHYPTAILYRPSLIAAAAVFAARCTLGRSPF-WTSTLKHYT 460
Query: 240 NYKASELKTTVLELIDLQLNTKRCRLNAVREKY 272
Y +L+ +++L +L AV +K+
Sbjct: 461 GYSEEQLRDCAKIMVNLHAAAPGSKLRAVYKKF 559
>AW756565 similar to PIR|T07675|T07 cyclin a2-type mitosis-specific -
soybean, partial (28%)
Length = 472
Score = 94.0 bits (232), Expect = 7e-20
Identities = 56/140 (40%), Positives = 87/140 (62%), Gaps = 2/140 (1%)
Frame = +2
Query: 16 IVDIDSKLRDSPIWTSYAPDIYTNIHVRECERRPLANYMETLQQDITPGMRGILVDWLVE 75
IV+ID+ D+ + +Y DIY ++ ++RP ++M+T+Q+DI MR ILVDWLVE
Sbjct: 53 IVNIDNIYSDTQLCATYVCDIYKHLRES*EKKRPSTDFMDTIQKDINVSMRTILVDWLVE 232
Query: 76 VADEFKLVPRHTLPCCESH--*SIPFPTFDYQTKAAVAWYYLHADFIYEEICAPGVEDFC 133
VA+E++LVP TL ++ S+ + + Q + + YEEICAP VE++
Sbjct: 233 VAEEYRLVP-ETLYLTVNYLDRSLSWNAMNRQRIQLLGVSCMMIASKYEEICAPHVEEYR 409
Query: 134 VITDNTYSRQEVLKMEKEVL 153
ITD+TY ++EVL+ME V+
Sbjct: 410 YITDDTYLKEEVLQMESCVV 469
>CO979059
Length = 766
Score = 90.5 bits (223), Expect = 8e-19
Identities = 47/110 (42%), Positives = 69/110 (62%), Gaps = 1/110 (0%)
Frame = -2
Query: 170 RRFIQVAQTS-SKVAQADLEFLANYLAELALVEYSFLQFQPSKIAASSVLLARWTLNQSE 228
RRF++ A ++ LE+L N++AEL+L+EYS L + PS IAAS + LAR+ L S+
Sbjct: 765 RRFVRAAAHDVQEIXSLQLEYLTNFIAELSLLEYSMLSYPPSLIAASVIFLARFILFPSK 586
Query: 229 HPWNPTLEHYTNYKASELKTTVLELIDLQLNTKRCRLNAVREKYQHQKVR 278
PWN TL+HYT Y+ S+L V +L L ++ L A+R+KY K +
Sbjct: 585 KPWNSTLQHYTLYRPSDLCACVKDLHRLCCSSHDSNLPAIRDKYSQHKYK 436
>BM188658 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (32%)
Length = 438
Score = 77.4 bits (189), Expect = 7e-15
Identities = 47/111 (42%), Positives = 71/111 (63%), Gaps = 3/111 (2%)
Frame = +3
Query: 46 ERRPLANYMETLQQDITPGMRGILVDWLVEVADEFKLVPRHTLPCCESH*SIPFPTFDYQ 105
+RRP+ +YM+ +Q+ +T MR ILVDWLVEVA+E+KL+ TL S+ F + +
Sbjct: 108 KRRPMIDYMDKVQKQVTTTMRTILVDWLVEVAEEYKLL-SDTLHLSVSYID-RFLSVNPV 281
Query: 106 TKAAVAWYYLHADFI---YEEICAPGVEDFCVITDNTYSRQEVLKMEKEVL 153
+K+ + + + I YEE+ P V+ FC ITDNTY + EV+KME ++L
Sbjct: 282 SKSRLQLLGVSSMLIAAKYEEVDPPRVDPFCNITDNTYHKAEVVKMEADML 434
>TC218840 similar to UP|CG1B_MEDVA (P46277) G2/mitotic-specific cyclin 1
(B-like cyclin) (CycMs1), partial (32%)
Length = 922
Score = 73.2 bits (178), Expect = 1e-13
Identities = 44/124 (35%), Positives = 65/124 (51%)
Frame = +1
Query: 153 LNLLHFQLAVPTIKTFLRRFIQVAQTSSKVAQADLEFLANYLAELALVEYSFLQFQPSKI 212
+NLL F ++VPT F++RF++ AQ K LE LA +L EL LVEY L+F PS +
Sbjct: 1 VNLLQFNMSVPTAYVFMKRFLKAAQADRK-----LELLAFFLVELTLVEYEMLKFPPSLL 165
Query: 213 AASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTKRCRLNAVREKY 272
AA++V A+ T+ + W+ T E ++NY +L + +L V KY
Sbjct: 166 AAAAVYTAQCTIYGFKQ-WSKTCEWHSNYSEDQLLECSTLMAAFHQKAGNGKLTGVHRKY 342
Query: 273 QHQK 276
K
Sbjct: 343 CSSK 354
>TC232525 homologue to UP|Q39880 (Q39880) Mitotic cyclin b1-type, partial
(73%)
Length = 1242
Score = 63.5 bits (153), Expect = 1e-10
Identities = 53/148 (35%), Positives = 76/148 (50%), Gaps = 5/148 (3%)
Frame = +1
Query: 16 IVDIDSKLRDSPIWTS-YAPDIYTNIHVRECERRPLANYMETLQQDITPGMRGILVDWLV 74
+++ID+ D+ + + Y DIY E E + +YM + Q DI MR ILVDWL+
Sbjct: 763 VMNIDATDMDNELAAAEYIDDIYKFYKETE-EEGCVHDYMGS-QPDINAKMRSILVDWLI 936
Query: 75 EVADEFKLVPRH---TLPCCESH*SIP-FPTFDYQTKAAVAWYYLHADFIYEEICAPGVE 130
EV +F+L+P TL + S+ P + Q + YEEI AP V
Sbjct: 937 EVHRKFELMPETLYLTLNIVDRFLSVKAVPRRELQLVGISSMLIASK---YEEIWAPEVN 1107
Query: 131 DFCVITDNTYSRQEVLKMEKEVLNLLHF 158
DF I+DN Y Q+VL MEK++L L +
Sbjct: 1108DFECISDNAYVSQQVLMMEKKILRKLEW 1191
>BQ453200 similar to GP|7242793|emb| cyclin A3.1 {Pisum sativum}, partial
(18%)
Length = 401
Score = 60.8 bits (146), Expect = 7e-10
Identities = 33/65 (50%), Positives = 40/65 (60%), Gaps = 1/65 (1%)
Frame = +2
Query: 213 AASSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTKRCRLNAVREKY 272
AAS V LAR+ + HPWN L T YK ++LK VL L DL L+ + L AVREKY
Sbjct: 2 AASVVFLARFMFSTKTHPWNLALHQLTRYKPADLKECVLNLHDLYLSRRGASLQAVREKY 181
Query: 273 -QHQK 276
QH+K
Sbjct: 182 KQHKK 196
>TC209822 similar to UP|Q9M4K9 (Q9M4K9) Cyclin A3.1, partial (24%)
Length = 640
Score = 58.5 bits (140), Expect = 3e-09
Identities = 29/64 (45%), Positives = 37/64 (57%)
Frame = +2
Query: 215 SSVLLARWTLNQSEHPWNPTLEHYTNYKASELKTTVLELIDLQLNTKRCRLNAVREKYQH 274
S V LAR+ + HPWN L T YK ++LK VL L DL L+ + L AVREKY+
Sbjct: 2 SVVFLARFMFSTKTHPWNSALHQLTRYKPADLKECVLNLHDLYLSRRGASLQAVREKYKQ 181
Query: 275 QKVR 278
K +
Sbjct: 182 HKFK 193
>AW705385 similar to PIR|T07672|T076 cyclin a2-type mitosis-specific -
soybean, partial (20%)
Length = 428
Score = 57.8 bits (138), Expect = 6e-09
Identities = 30/61 (49%), Positives = 37/61 (60%), Gaps = 6/61 (9%)
Frame = +1
Query: 44 ECERRPLANYMETLQQDITPGMRGILVDWLVEVAD------EFKLVPRHTLPCCESH*SI 97
E +R+PL NYME LQ+DI P MR ILVDWLVE D + L+ H P +H*+
Sbjct: 193 ELQRKPLTNYMEKLQKDINPSMRRILVDWLVECDDYKVKQEQIPLLTSHVPPSSTAH*TS 372
Query: 98 P 98
P
Sbjct: 373 P 375
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,566,267
Number of Sequences: 63676
Number of extensions: 195050
Number of successful extensions: 1330
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1318
length of query: 293
length of database: 12,639,632
effective HSP length: 97
effective length of query: 196
effective length of database: 6,463,060
effective search space: 1266759760
effective search space used: 1266759760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146722.7


