

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.4 - phase: 0 /pseudo
(471 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
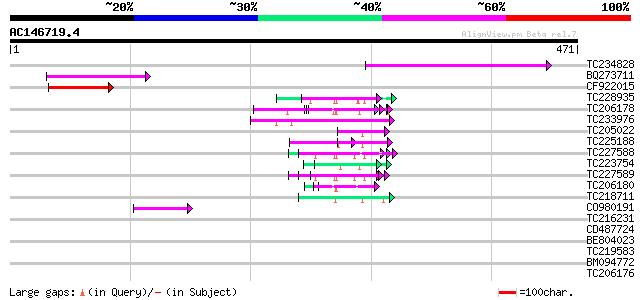
Score E
Sequences producing significant alignments: (bits) Value
TC234828 121 8e-28
BQ273711 87 1e-17
CF922015 85 6e-17
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 55 5e-08
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 55 9e-08
TC233976 52 4e-07
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 50 2e-06
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 50 2e-06
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 49 4e-06
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 48 1e-05
TC227589 48 1e-05
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 47 1e-05
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 46 4e-05
CO980191 42 6e-04
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 40 0.002
CD487724 40 0.003
BE804023 36 0.032
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 36 0.042
BM094772 33 0.36
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 29 4.0
>TC234828
Length = 857
Score = 121 bits (303), Expect = 8e-28
Identities = 61/155 (39%), Positives = 87/155 (55%)
Frame = +3
Query: 296 NCNEEGHISSQCTQPKKVRTGGKVFALTGTQTVNEDRLIRGTCFFNSTHLIAIIDTGATH 355
N G++S+ + + +VFA++G++ D LIRG C L + D+GATH
Sbjct: 66 NSTRSGNVSNNNISGGRPKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATH 245
Query: 356 CFIVVDCAYKLGLIVSDMNGEMVVETPAKGSVTTSLVCLSCLLSMFGRDFEVDLVCLPLS 415
FI C +LGL +++ +MVV TP VTTS VCL C + + GR F DL+CLPL+
Sbjct: 246 SFISHACVERLGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLA 425
Query: 416 GVDVIFGMNWLEYNHFHINCFSKTVHFSSAEEESE 450
+DVI GM+WL NH ++C K + F SE
Sbjct: 426 HLDVILGMDWLSTNHIFLDCKEKMLVFGGDVVPSE 530
>BQ273711
Length = 409
Score = 87.4 bits (215), Expect = 1e-17
Identities = 39/87 (44%), Positives = 50/87 (56%)
Frame = -2
Query: 31 RMLETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEADDWW 90
R L F +NHPP F G YDP+GA+ WL E E+IF M C E KV + T ML EA++WW
Sbjct: 261 RGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYATFMLQGEAENWW 82
Query: 91 VSLLPILEQDGAAVTWVVFRREFLNMY 117
+ P G + W F+ +FL Y
Sbjct: 81 KFVKPSFVAPGGVIPWNAFKDKFLENY 1
>CF922015
Length = 172
Score = 85.1 bits (209), Expect = 6e-17
Identities = 36/54 (66%), Positives = 46/54 (84%)
Frame = -3
Query: 33 LETFLRNHPPTFKGRYDPDGAQKWLKEVERIFRVMQCSEVQKVRFGTHMLAEEA 86
L+ F RN+PPTFKG YDP+GA+ WL+E+E+IFRVM+C + QKV F THMLA+EA
Sbjct: 164 LDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 55.5 bits (132), Expect = 5e-08
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 13/80 (16%)
Frame = +1
Query: 243 PKRRDAPTEIICFKCGEKGHKSNVCD------RDEKKCFRCGQKGHTLADC----KRGDV 292
P++ + IC +C +GH++ C +D K C+ CG+ GH L C + G
Sbjct: 313 PEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQEGGT 492
Query: 293 V---CYNCNEEGHISSQCTQ 309
C+ CN+ GH+S C Q
Sbjct: 493 KFAECFVCNQRGHLSKNCPQ 552
Score = 49.3 bits (116), Expect = 4e-06
Identities = 36/127 (28%), Positives = 49/127 (38%), Gaps = 27/127 (21%)
Frame = +1
Query: 222 RPKPYSAPTDKGKQRLNDERRPKRRDA----------------PTEIICFKCGEKGHKSN 265
+P P P K K+ N + KR + P E CF C H +
Sbjct: 130 KPTPPKDPDKKKKKNKNSAFKRKRPEPKPGSRKRHLLRVPGMKPGES-CFICKAMDHIAK 306
Query: 266 VCDRD-----EKKCFRCGQKGHTLADCK------RGDVVCYNCNEEGHISSQCTQPKKVR 314
+C K C RC ++GH +C + CYNC E GH +QC P ++
Sbjct: 307 LCPEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHP--LQ 480
Query: 315 TGGKVFA 321
GG FA
Sbjct: 481 EGGTKFA 501
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 54.7 bits (130), Expect = 9e-08
Identities = 33/118 (27%), Positives = 51/118 (42%), Gaps = 2/118 (1%)
Frame = +2
Query: 203 DTKAHYKVMSERRGKGQ*SRPKPYSAP--TDKGKQRLNDERRPKRRDAPTEIICFKCGEK 260
+ K +K+ S+ R + + P +D+ R RR RR + +C C
Sbjct: 185 ERKKKWKMSSDSRSRSRSRSRSPMDRKIRSDRFSYRDAPYRRDSRRGFSRDNLCKNCKRP 364
Query: 261 GHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKKVRTGGK 318
GH + C + C CG GH ++C + C+NC E GH++S C T GK
Sbjct: 365 GHYARECP-NVAICHNCGLPGHIASECTTKSL-CWNCKEPGHMASSCPNEGICHTCGK 532
Score = 54.7 bits (130), Expect = 9e-08
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Frame = +2
Query: 247 DAPTEIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKR-----GDV-VCYNCNEE 300
+ T+ +C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC ++
Sbjct: 437 ECTTKSLCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQ 613
Query: 301 GHISSQCTQPK 311
GHI+++CT K
Sbjct: 614 GHIAAECTNEK 646
Score = 48.9 bits (115), Expect = 5e-06
Identities = 28/96 (29%), Positives = 39/96 (40%), Gaps = 27/96 (28%)
Frame = +2
Query: 249 PTEIICFKCGEKGHKSNVCDR-------------------------DEKKCFRCGQKGHT 283
P E IC CG+ GH++ C +EK C C + GH
Sbjct: 500 PNEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEKACNNCRKTGHL 679
Query: 284 LADCKRGDVVCYNCNEEGHISSQCTQPKKV--RTGG 317
DC D +C CN GH++ QC + + R+GG
Sbjct: 680 ARDCP-NDPICNLCNVSGHVARQCPKANVLGDRSGG 784
Score = 43.1 bits (100), Expect = 3e-04
Identities = 24/87 (27%), Positives = 33/87 (37%), Gaps = 25/87 (28%)
Frame = +2
Query: 246 RDAPTEIICFKCGEKGHKSNVC-------DRD------------------EKKCFRCGQK 280
RD P + IC C GH + C DR + C C Q
Sbjct: 683 RDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVVCRNCQQL 862
Query: 281 GHTLADCKRGDVVCYNCNEEGHISSQC 307
GH DC ++C+NC GH++ +C
Sbjct: 863 GHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 35.0 bits (79), Expect = 0.072
Identities = 13/40 (32%), Positives = 19/40 (47%)
Frame = +2
Query: 251 EIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRG 290
+++C C + GH S C C CG +GH +C G
Sbjct: 833 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 952
>TC233976
Length = 763
Score = 52.4 bits (124), Expect = 4e-07
Identities = 32/128 (25%), Positives = 54/128 (42%), Gaps = 9/128 (7%)
Frame = +1
Query: 201 EEDTKAHYKVMSERRGKGQ*----SRPKPYSAPTDK-----GKQRLNDERRPKRRDAPTE 251
+++ A YK + GK + SR KPYSAP + G++
Sbjct: 25 QQEKVAFYKNANASHGKEKKPMTHSRAKPYSAPLEYENHYGGQRTSGGHHLAGGSSQLVN 204
Query: 252 IICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPK 311
+ G G + +C +CG+ GH +C +V C+N +GH+S+ C P+
Sbjct: 205 RVSQPAGRGGSGAPAIVTTPLRCRKCGRLGHNAHECTYREVTCFNYQGKGHLSTNCPHPR 384
Query: 312 KVRTGGKV 319
K + G +
Sbjct: 385 KEKRSGSL 408
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 50.1 bits (118), Expect = 2e-06
Identities = 23/46 (50%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Frame = +2
Query: 273 KCFRCGQKGHTLADCKRGD--VVCYNCNEEGHISSQC-TQPKKVRT 315
+CF CG GH DCK GD CY C E GHI C PKK+ T
Sbjct: 311 RCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSPKKLST 448
Score = 41.2 bits (95), Expect = 0.001
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDRDE--KKCFRCGQKGHTLADCK 288
CF CG GH + C + KC+RCG++GH +CK
Sbjct: 314 CFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCK 424
Score = 28.1 bits (61), Expect = 8.8
Identities = 9/20 (45%), Positives = 12/20 (60%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDRDEKK 273
C++CGE+GH C KK
Sbjct: 380 CYRCGERGHIERNCKNSPKK 439
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 50.1 bits (118), Expect = 2e-06
Identities = 23/49 (46%), Positives = 26/49 (52%), Gaps = 3/49 (6%)
Frame = +1
Query: 273 KCFRCGQKGHTLADCKRGD--VVCYNCNEEGHISSQC-TQPKKVRTGGK 318
+CF CG GH DCK GD CY C E GHI C PKK+ G+
Sbjct: 361 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKLSQRGR 507
Score = 41.6 bits (96), Expect = 8e-04
Identities = 20/58 (34%), Positives = 27/58 (46%), Gaps = 2/58 (3%)
Frame = +1
Query: 233 GKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDRDE--KKCFRCGQKGHTLADCK 288
G + D R P CF CG GH + C + KC+RCG++GH +CK
Sbjct: 301 GPRGSRDREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 474
Score = 29.6 bits (65), Expect = 3.0
Identities = 10/27 (37%), Positives = 16/27 (59%)
Frame = +1
Query: 254 CFKCGEKGHKSNVCDRDEKKCFRCGQK 280
C++CGE+GH C KK + G++
Sbjct: 430 CYRCGERGHIEKNCKNSPKKLSQRGRR 510
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 49.3 bits (116), Expect = 4e-06
Identities = 30/85 (35%), Positives = 38/85 (44%), Gaps = 13/85 (15%)
Frame = +1
Query: 241 RRPKRRDAPTEIICFKCGEKGHKSNVCDRD----EKKCFRCGQKGHTLADCKR-GDVV-- 293
R +D EI C+ C GH V D E C++CGQ GHT C R D +
Sbjct: 307 RNDYSQDDLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITS 486
Query: 294 ------CYNCNEEGHISSQCTQPKK 312
C+ C EEGH + +CT K
Sbjct: 487 GATPSSCFKCGEEGHFARECTSSIK 561
Score = 48.9 bits (115), Expect = 5e-06
Identities = 28/97 (28%), Positives = 39/97 (39%), Gaps = 11/97 (11%)
Frame = +1
Query: 232 KGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDRD-------EKKCFRCGQKGHTL 284
KG R D + + IC KCG GH C D E +C+ C + GH
Sbjct: 199 KGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSCRNDYSQDDLKEIQCYVCKRLGHLC 378
Query: 285 A----DCKRGDVVCYNCNEEGHISSQCTQPKKVRTGG 317
D G++ CY C + GH C++ + T G
Sbjct: 379 CVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITSG 489
Score = 48.1 bits (113), Expect = 8e-06
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 4/86 (4%)
Frame = +1
Query: 241 RRPKRRDAPTEI--ICFKCGEKGHKSNVCD--RDEKKCFRCGQKGHTLADCKRGDVVCYN 296
R P+ D P CF CGE+GH + C + +K C+ CG GH C + C+
Sbjct: 16 RGPRYFDPPDNSWGACFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSKVQ-DCFI 192
Query: 297 CNEEGHISSQCTQPKKVRTGGKVFAL 322
C + GH + C P+K + K A+
Sbjct: 193 CKKGGHRAKDC--PEKHTSTSKSIAI 264
Score = 31.2 bits (69), Expect = 1.0
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = +1
Query: 254 CFKCGEKGHKSNVCDRDEKKCFRCGQKGHT 283
CFKCGE+GH + C K R + HT
Sbjct: 505 CFKCGEEGHFARECTSSIKSGKRNWESSHT 594
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/95 (29%), Positives = 36/95 (37%), Gaps = 22/95 (23%)
Frame = +2
Query: 245 RRDAPTEIICFKCGEKGHKSNVCDRDEKK----------CFRCGQKGHTLADCKR----- 289
RR+ C++CGE GH + C+R CF CG GH DC R
Sbjct: 17 RRNGGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGS 196
Query: 290 -------GDVVCYNCNEEGHISSQCTQPKKVRTGG 317
G C+ C GH++ C K GG
Sbjct: 197 VGIGGGGGGGSCFRCGGFGHMARDCATGKGNIGGG 301
Score = 47.4 bits (111), Expect = 1e-05
Identities = 23/80 (28%), Positives = 32/80 (39%), Gaps = 24/80 (30%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDRDEKK---------CFRCGQKGHTLADCKR--------------- 289
CF+CG GH + C + CFRCG+ GH DC
Sbjct: 230 CFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFGGGGGSGGGG 409
Query: 290 GDVVCYNCNEEGHISSQCTQ 309
G C+NC + GH + +C +
Sbjct: 410 GKSTCFNCGKPGHFARECVE 469
>TC227589
Length = 547
Score = 47.8 bits (112), Expect = 1e-05
Identities = 25/79 (31%), Positives = 36/79 (44%), Gaps = 4/79 (5%)
Frame = +2
Query: 241 RRPKRRDAPTEI--ICFKCGEKGHKSNVCD--RDEKKCFRCGQKGHTLADCKRGDVVCYN 296
R P+ D P CF CGE GH + C + +K C+ CG GH C + C+
Sbjct: 2 RGPRYFDPPDSSWGACFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTKAQ-DCFI 178
Query: 297 CNEEGHISSQCTQPKKVRT 315
C + GH + C + R+
Sbjct: 179 CKKGGHRAKDCLEKHTSRS 235
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/89 (29%), Positives = 36/89 (40%), Gaps = 11/89 (12%)
Frame = +2
Query: 232 KGKQRLNDERRPKRRDAPTEIICFKCGEKGHKSNVCDRD-------EKKCFRCGQKGHTL 284
KG R D + + IC KCG GH C D E +C+ C + GH
Sbjct: 185 KGGHRAKDCLEKHTSRSKSVAICLKCGNSGHDMFSCRNDYSPDDLKEIQCYVCKRVGHLC 364
Query: 285 A----DCKRGDVVCYNCNEEGHISSQCTQ 309
D G++ CY C + GH C++
Sbjct: 365 CVNTDDATPGEISCYKCGQLGHTGLACSK 451
Score = 47.0 bits (110), Expect = 2e-05
Identities = 28/73 (38%), Positives = 34/73 (46%), Gaps = 13/73 (17%)
Frame = +2
Query: 251 EIICFKCGEKGHKSNVCDRD----EKKCFRCGQKGHT-LADCKRGDVV--------CYNC 297
EI C+ C GH V D E C++CGQ GHT LA K D + C C
Sbjct: 323 EIQCYVCKRVGHLCCVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCCKC 502
Query: 298 NEEGHISSQCTQP 310
E GH + +CT P
Sbjct: 503 GEAGHFAQECTSP 541
Score = 39.7 bits (91), Expect = 0.003
Identities = 18/48 (37%), Positives = 24/48 (49%), Gaps = 9/48 (18%)
Frame = +2
Query: 249 PTEIICFKCGEKGHKSNVCDR--DE-------KKCFRCGQKGHTLADC 287
P EI C+KCG+ GH C + DE C +CG+ GH +C
Sbjct: 389 PGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCCKCGEAGHFAQEC 532
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 47.4 bits (111), Expect = 1e-05
Identities = 21/55 (38%), Positives = 30/55 (54%)
Frame = +3
Query: 253 ICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQC 307
+C C ++GH + C +EK C C + GH DC D +C CN GH++ QC
Sbjct: 75 LCNNCYKQGHIAAECT-NEKACNNCRKTGHLARDCP-NDPICNLCNVSGHVARQC 233
Score = 43.9 bits (102), Expect = 2e-04
Identities = 22/84 (26%), Positives = 32/84 (37%), Gaps = 22/84 (26%)
Frame = +3
Query: 246 RDAPTEIICFKCGEKGHKSNVCDRD----------------------EKKCFRCGQKGHT 283
RD P + IC C GH + C + + C C Q GH
Sbjct: 168 RDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDVVCRNCQQLGHM 347
Query: 284 LADCKRGDVVCYNCNEEGHISSQC 307
DC ++C+NC GH++ +C
Sbjct: 348 SRDCMGPLMICHNCGGRGHLAYEC 419
Score = 42.7 bits (99), Expect = 3e-04
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 6/57 (10%)
Frame = +3
Query: 257 CGEKGHKSNVCDR------DEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQC 307
CG+ GH++ C D + C C ++GH A+C + C NC + GH++ C
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGHLARDC 176
Score = 37.4 bits (85), Expect = 0.015
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 6/41 (14%)
Frame = +3
Query: 277 CGQKGHTLADCKR-----GDV-VCYNCNEEGHISSQCTQPK 311
CG+ GH +C GD+ +C NC ++GHI+++CT K
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEK 131
Score = 35.0 bits (79), Expect = 0.072
Identities = 13/40 (32%), Positives = 19/40 (47%)
Frame = +3
Query: 251 EIICFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRG 290
+++C C + GH S C C CG +GH +C G
Sbjct: 309 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 428
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 45.8 bits (107), Expect = 4e-05
Identities = 27/97 (27%), Positives = 38/97 (38%), Gaps = 18/97 (18%)
Frame = -3
Query: 241 RRPKRRDAPTEIICFKCGEKGHKSNVCDR------DEKKCFRCGQKGHTLADCKRGD--- 291
+ P + + + C C + GH+ C R DE+ CF CG+ GH+L C
Sbjct: 560 KSPFKHHGESSLRCRACRQPGHRFQQCQRLKCLSMDEEVCFFCGEIGHSLGKCDVSQAGG 381
Query: 292 ---VVCYNCNEEGHISSQCTQ------PKKVRTGGKV 319
C C GH S C Q PK + G +
Sbjct: 380 GRFAKCLLCYGHGHFSYNCPQNGHGIDPKVLAVNGAI 270
>CO980191
Length = 802
Score = 42.0 bits (97), Expect = 6e-04
Identities = 21/49 (42%), Positives = 28/49 (56%)
Frame = +1
Query: 104 VTWVVFRREFLNMYFPEDVRGKKEIEFLEMKQGDMSVTEYAAKFVELAK 152
+T VF+ FL + ED+ K+ EFLE+K G+M V Y F EL K
Sbjct: 364 IT*EVFKEIFLEKHLHEDI*NHKKTEFLELKHGNMIVANYLVNFDELPK 510
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/54 (37%), Positives = 24/54 (44%)
Frame = +3
Query: 254 CFKCGEKGHKSNVCDRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQC 307
C+KCGE GH + C + R G G G CYNC E GH + C
Sbjct: 231 CYKCGETGHIARDCSQGGGGGGRYGGGG-------GGGGSCYNCGESGHFARDC 371
Score = 28.5 bits (62), Expect = 6.8
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +3
Query: 294 CYNCNEEGHISSQCTQ 309
CY C E GHI+ C+Q
Sbjct: 231 CYKCGETGHIARDCSQ 278
>CD487724
Length = 676
Score = 39.7 bits (91), Expect = 0.003
Identities = 32/129 (24%), Positives = 57/129 (43%), Gaps = 9/129 (6%)
Frame = +3
Query: 345 LIAIIDTGATHCFIVVDCAYKLGLIVSDMNGEMVVETPAKGSVTTSLVCLSCLLSMFGRD 404
L+ ++D G+TH F+ +LGL V+ T+ +C + +S+ +
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTT-ICEAIPISIQNIE 200
Query: 405 FEVDLVCLPLSGVDVIFGMNWL--------EYNHFHINCF-SKTVHFSSAEEESEAKFLT 455
F V L LP+ G +++ G+ WL +YN + F + E E++ L
Sbjct: 201 FLVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLLN 380
Query: 456 TKQLKQLEQ 464
QL++L Q
Sbjct: 381 HHQLRRLHQ 407
>BE804023
Length = 407
Score = 36.2 bits (82), Expect = 0.032
Identities = 19/60 (31%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Frame = -3
Query: 254 CFKCGEKGHKSNVC-DRDEKKCFRCGQKGHTLADCKRGDVVCYNCNEEGHISSQCTQPKK 312
C C +KGH C R + KC +C Q GH + V+C N N+ + +++ PK+
Sbjct: 219 C*HCRKKGHPPFRC*KRPDAKCSQCNQMGHEV-------VICKNRNQSHNEAAKVVDPKE 61
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 35.8 bits (81), Expect = 0.042
Identities = 16/55 (29%), Positives = 22/55 (39%), Gaps = 20/55 (36%)
Frame = +2
Query: 273 KCFRCGQKGHTLADCKRGDVV--------------------CYNCNEEGHISSQC 307
+C+ CG+ GH DC G C+NC EEGH + +C
Sbjct: 422 ECYNCGRIGHLARDCYHGQGGGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFAREC 586
>BM094772
Length = 421
Score = 32.7 bits (73), Expect = 0.36
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 6/36 (16%)
Frame = +2
Query: 254 CFKCGEKGHKSNVCDR------DEKKCFRCGQKGHT 283
C C + GH+ C R DE+ CF CG+ GH+
Sbjct: 314 CRACRQPGHRFQQCQRLKCLSMDEEVCFFCGEIGHS 421
Score = 30.0 bits (66), Expect = 2.3
Identities = 13/37 (35%), Positives = 18/37 (48%), Gaps = 6/37 (16%)
Frame = +2
Query: 272 KKCFRCGQKGHTLADCKR------GDVVCYNCNEEGH 302
++C C Q GH C+R + VC+ C E GH
Sbjct: 308 QRCRACRQPGHRFQQCQRLKCLSMDEEVCFFCGEIGH 418
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 29.3 bits (64), Expect = 4.0
Identities = 9/19 (47%), Positives = 12/19 (62%)
Frame = +3
Query: 290 GDVVCYNCNEEGHISSQCT 308
G CYNC E GH++ C+
Sbjct: 423 GGGACYNCGESGHLARDCS 479
Score = 29.3 bits (64), Expect = 4.0
Identities = 15/66 (22%), Positives = 19/66 (28%), Gaps = 32/66 (48%)
Frame = +3
Query: 274 CFRCGQKGHTLADCK--------------------------------RGDVVCYNCNEEG 301
C+ CG+ GH DC G CY+C E G
Sbjct: 435 CYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGGGGGRYVGGGGGGGGGGSCYSCGESG 614
Query: 302 HISSQC 307
H + C
Sbjct: 615 HFARDC 632
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,279,859
Number of Sequences: 63676
Number of extensions: 364216
Number of successful extensions: 1789
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1611
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1715
length of query: 471
length of database: 12,639,632
effective HSP length: 101
effective length of query: 370
effective length of database: 6,208,356
effective search space: 2297091720
effective search space used: 2297091720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146719.4


