

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.10 + phase: 0 /pseudo
(112 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
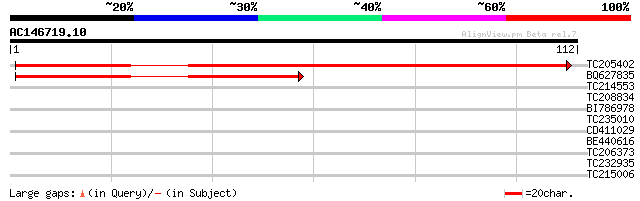
Score E
Sequences producing significant alignments: (bits) Value
TC205402 similar to UP|Q9SIV2 (Q9SIV2) 26S proteasome regulatory... 156 1e-39
BQ627835 homologue to GP|20198043|gb 26S proteasome regulatory s... 64 1e-11
TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 27 1.9
TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I... 26 3.2
BI786978 25 4.2
TC235010 homologue to GB|AAD04176.1|4139230|AF109243 mismatch re... 25 5.5
CD411029 similar to SP|Q9LK09|SY32 Syntaxin 32 (AtSYP32). [Mouse... 25 7.2
BE440616 25 7.2
TC206373 similar to UP|Q8L7S0 (Q8L7S0) At1g09730/F21M12_12, part... 25 7.2
TC232935 24 9.4
TC215006 similar to UP|RL18_CICAR (O65729) 60S ribosomal protein... 24 9.4
>TC205402 similar to UP|Q9SIV2 (Q9SIV2) 26S proteasome regulatory subunit S2
(RPN1) (26S proteasome subunit RPN1a), partial (51%)
Length = 1624
Score = 156 bits (395), Expect = 1e-39
Identities = 83/110 (75%), Positives = 90/110 (81%)
Frame = +1
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDV 61
L++ ++ GKYHYVLYFLVLAMQ P MLLTV ENLKPL VP+RVGQAVDV
Sbjct: 1048 LDMKAIVLGKYHYVLYFLVLAMQ-----------PRMLLTVDENLKPLSVPVRVGQAVDV 1194
Query: 62 VGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
VGQAGRPK ITGFQTHST VLLAAGDRAELATEKYIP+SP+LE FVILKE
Sbjct: 1195 VGQAGRPKTITGFQTHSTPVLLAAGDRAELATEKYIPLSPILEGFVILKE 1344
>BQ627835 homologue to GP|20198043|gb 26S proteasome regulatory subunit S2
(RPN1) {Arabidopsis thaliana}, partial (12%)
Length = 423
Score = 63.5 bits (153), Expect = 1e-11
Identities = 35/57 (61%), Positives = 40/57 (69%)
Frame = +3
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQA 58
L++ ++ GKYHYVLYFLVLAMQ P MLLTV ENLKPL VP+RVGQA
Sbjct: 285 LDMKAIVLGKYHYVLYFLVLAMQ-----------PRMLLTVDENLKPLSVPVRVGQA 422
>TC214553 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (52%)
Length = 1808
Score = 26.6 bits (57), Expect = 1.9
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 7/49 (14%)
Frame = +1
Query: 67 RPKKITGFQTHSTS----VLLAAGDRAELATEK---YIPISPVLEAFVI 108
+P+++ G HS S +LL + L K +IP+ P+ AFVI
Sbjct: 664 QPERVIGINPHSDSGALTILLQVNEVEGLQIRKDGKWIPVKPLSNAFVI 810
>TC208834 similar to UP|Q39224 (Q39224) SRG1 protein (F6I1.30/F6I1.30)
(At1g17020/F6I1.30), partial (54%)
Length = 1070
Score = 25.8 bits (55), Expect = 3.2
Identities = 13/48 (27%), Positives = 25/48 (52%), Gaps = 7/48 (14%)
Frame = +2
Query: 68 PKKITGFQTHST----SVLLAAGDRAELATEK---YIPISPVLEAFVI 108
P+K+ G HS ++LL + L K ++P+ P++ AF++
Sbjct: 479 PEKVIGLTPHSDGIGLAILLQLNEVEGLQIRKDGLWVPVKPLINAFIV 622
>BI786978
Length = 431
Score = 25.4 bits (54), Expect = 4.2
Identities = 9/30 (30%), Positives = 18/30 (60%)
Frame = +1
Query: 12 YHYVLYFLVLAMQLTLGYYILLQKPTMLLT 41
+H +LY++ L + + L YYI + ++ T
Sbjct: 340 FHELLYYIKLILDILLNYYISNMRVAIIYT 429
>TC235010 homologue to GB|AAD04176.1|4139230|AF109243 mismatch repair protein
{Arabidopsis thaliana;} , partial (7%)
Length = 760
Score = 25.0 bits (53), Expect = 5.5
Identities = 16/36 (44%), Positives = 22/36 (60%)
Frame = -2
Query: 14 YVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPL 49
Y+L L+LA +L + L K MLL +GE L+PL
Sbjct: 399 YLLTSLLLA-KLAFCPWFLEAKQQMLLEIGEPLQPL 295
>CD411029 similar to SP|Q9LK09|SY32 Syntaxin 32 (AtSYP32). [Mouse-ear cress]
{Arabidopsis thaliana}, partial (50%)
Length = 671
Score = 24.6 bits (52), Expect = 7.2
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +2
Query: 10 GKYHYVLYFLVLAMQLTLGYYILLQK 35
G Y +L L+L LTL Y+LL+K
Sbjct: 362 GNYLLLLLLLLLQQWLTLPIYLLLRK 439
>BE440616
Length = 450
Score = 24.6 bits (52), Expect = 7.2
Identities = 10/20 (50%), Positives = 12/20 (60%)
Frame = +2
Query: 12 YHYVLYFLVLAMQLTLGYYI 31
YH LYF L M +GYY+
Sbjct: 218 YHTPLYFSYL*MYFLVGYYV 277
>TC206373 similar to UP|Q8L7S0 (Q8L7S0) At1g09730/F21M12_12, partial (18%)
Length = 1651
Score = 24.6 bits (52), Expect = 7.2
Identities = 11/27 (40%), Positives = 18/27 (65%)
Frame = -2
Query: 13 HYVLYFLVLAMQLTLGYYILLQKPTML 39
H+VL+ ++AM L +++L QKP L
Sbjct: 1236 HHVLHQKLVAMVLEFPHHLLHQKPAAL 1156
>TC232935
Length = 1091
Score = 24.3 bits (51), Expect = 9.4
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +1
Query: 17 YFLVLAMQLTLGYYILL 33
YFLV+A+Q L +YIL+
Sbjct: 733 YFLVIAVQCFLEHYILI 783
>TC215006 similar to UP|RL18_CICAR (O65729) 60S ribosomal protein L18
(Fragment), complete
Length = 826
Score = 24.3 bits (51), Expect = 9.4
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +2
Query: 13 HYVLYFLVLAMQLTLGYYILLQK 35
HYVL+ VL + G+Y L K
Sbjct: 671 HYVLFSCVLEQFVNFGFYFSLSK 739
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.333 0.149 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,204,375
Number of Sequences: 63676
Number of extensions: 46282
Number of successful extensions: 294
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 290
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 292
length of query: 112
length of database: 12,639,632
effective HSP length: 88
effective length of query: 24
effective length of database: 7,036,144
effective search space: 168867456
effective search space used: 168867456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146719.10


