

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146711.3 - phase: 0
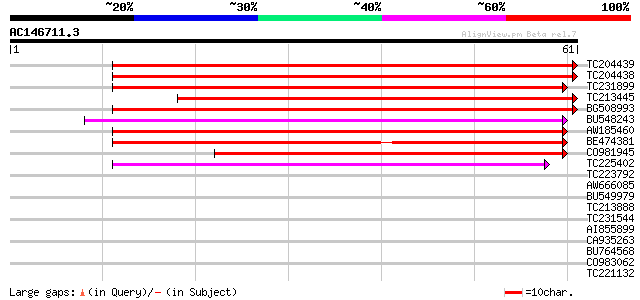
(61 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 57 2e-09
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 57 2e-09
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 52 5e-08
TC213445 51 1e-07
BG508993 49 7e-07
BU548243 46 4e-06
AW185460 45 8e-06
BE474381 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, pa... 42 8e-05
CO981945 39 5e-04
TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotr... 39 7e-04
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 38 0.001
AW666085 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, pa... 38 0.001
BU549979 38 0.001
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 37 0.002
TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, ... 37 0.003
AI855899 similar to GP|2244960|emb| retrotransposon like protein... 36 0.005
CA935263 35 0.006
BU764568 34 0.013
CO983062 33 0.023
TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON li... 33 0.030
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 57.0 bits (136), Expect = 2e-09
Identities = 27/50 (54%), Positives = 34/50 (68%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
DAD+AG +RKSTSG C +L L W +KQN ++LST EAEY +A S
Sbjct: 4273 DADWAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGS 4422
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 56.6 bits (135), Expect = 2e-09
Identities = 27/50 (54%), Positives = 34/50 (68%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
DAD+AG +RKSTSG C +L L W +KQN ++LST EAEY +A S
Sbjct: 4276 DADWAGSADDRKSTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGS 4425
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 52.4 bits (124), Expect = 5e-08
Identities = 24/49 (48%), Positives = 32/49 (64%)
Frame = +2
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
DAD+AG ++R+STSG C F+ L W +KQ +A S+ EAEY S A
Sbjct: 32 DADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMA 178
>TC213445
Length = 705
Score = 51.2 bits (121), Expect = 1e-07
Identities = 23/43 (53%), Positives = 30/43 (69%)
Frame = +1
Query: 19 KIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
K +R+STS +C F+ AL W +KQN++ LST EAEY SA S
Sbjct: 400 KTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARS 528
>BG508993
Length = 374
Score = 48.5 bits (114), Expect = 7e-07
Identities = 23/50 (46%), Positives = 32/50 (64%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
D+D+AGD +RKST+G F+ + WS +KQ + L T EAEY +A S
Sbjct: 70 DSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQGIVTLFTCEAEYVAATS 219
>BU548243
Length = 599
Score = 45.8 bits (107), Expect = 4e-06
Identities = 25/52 (48%), Positives = 30/52 (57%)
Frame = -1
Query: 9 SLLDADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
+L DA +A D + +ST GS FL L W RKQ A S+TEAEY S A
Sbjct: 596 ALCDAGWASDVDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIA 441
>AW185460
Length = 411
Score = 45.1 bits (105), Expect = 8e-06
Identities = 22/49 (44%), Positives = 30/49 (60%)
Frame = +2
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
D+D+AG + KSTSG L + W+ +KQ T+A ST EAEY + A
Sbjct: 260 DSDWAGSTDDMKSTSGYAFSLGSGMFSWASKKQATVAQSTAEAEYVAVA 406
>BE474381 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (20%)
Length = 406
Score = 41.6 bits (96), Expect = 8e-05
Identities = 22/49 (44%), Positives = 32/49 (64%)
Frame = +3
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
DAD+AG +R+STSG C F+ L S +KQ+ +A S+ EAE+ + A
Sbjct: 162 DADWAGSVTDRRSTSGYCTFVGGNLVS*S-KKQSVVARSSAEAEFRALA 305
>CO981945
Length = 584
Score = 38.9 bits (89), Expect = 5e-04
Identities = 19/38 (50%), Positives = 24/38 (63%)
Frame = -1
Query: 23 KSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
+STSG+C +L+Q W +KQ IA S EAEY S A
Sbjct: 440 RSTSGACIYLDQNRISW**KKQTLIACSIAEAEYHSLA 327
>TC225402 weakly similar to UP|Q6I923 (Q6I923) Copia-like retrotransposon
Hopscotch polyprotein, partial (7%)
Length = 1446
Score = 38.5 bits (88), Expect = 7e-04
Identities = 19/47 (40%), Positives = 28/47 (59%)
Frame = +2
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYES 58
DA++A I+R ST G C + + L W K N +A S+ EAEY++
Sbjct: 8 DANWAVSPIDRGSTLGYCVSIGENLVLWKSNK*NVVARSSAEAEYKA 148
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (7%)
Length = 804
Score = 38.1 bits (87), Expect = 0.001
Identities = 21/49 (42%), Positives = 28/49 (56%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
D+D+ D + KST G A S +KQ+ IALST EAEY +A+
Sbjct: 238 DSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAEYVAAS 384
>AW666085 similar to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial
(16%)
Length = 193
Score = 38.1 bits (87), Expect = 0.001
Identities = 20/49 (40%), Positives = 30/49 (60%)
Frame = +2
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
DAD+AG ++KSTSG C F+ L + Q+ +A+S+ AEY + A
Sbjct: 5 DADWAGSITDQKSTSGYCTFVWGNLVT*MNK*QDVVAMSSVYAEYRAMA 151
>BU549979
Length = 615
Score = 38.1 bits (87), Expect = 0.001
Identities = 19/45 (42%), Positives = 24/45 (53%)
Frame = -1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEY 56
D+D+AG R+STSG L + W KQ IA ST E E+
Sbjct: 477 DSDFAGCVDSRRSTSGYIFMLADGVVSWRSSKQTLIATSTMEVEF 343
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 37.0 bits (84), Expect = 0.002
Identities = 18/41 (43%), Positives = 24/41 (57%)
Frame = +3
Query: 21 ERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAAS 61
+RKST+G F+ W +KQ + LST EAEY +A S
Sbjct: 12 DRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATS 134
>TC231544 weakly similar to UP|Q9FLR2 (Q9FLR2) Polyprotein-like, partial
(16%)
Length = 662
Score = 36.6 bits (83), Expect = 0.003
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Frame = +3
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTI--ALSTTEAEYESAAS 61
DAD+A KS + C FL +L W +KQNT+ + S++EA+Y + S
Sbjct: 111 DADWATCIDSSKSITWYCFFLGSSLISWKAKKQNTVSRSSSSSEAKYRALTS 266
>AI855899 similar to GP|2244960|emb| retrotransposon like protein
{Arabidopsis thaliana}, partial (18%)
Length = 418
Score = 35.8 bits (81), Expect = 0.005
Identities = 16/33 (48%), Positives = 19/33 (57%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQ 44
D D+ D E +STSGSC F L WS +KQ
Sbjct: 319 DLDWGSDPAEMRSTSGSCIFSGSNLIAWSSKKQ 417
>CA935263
Length = 446
Score = 35.4 bits (80), Expect = 0.006
Identities = 16/40 (40%), Positives = 22/40 (55%)
Frame = +3
Query: 15 YAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEA 54
+ G +STSG C + + L W +KQNT A S+ EA
Sbjct: 3 WEGSPSNSRSTSGYCVLIGENLISWRSKKQNTFARSSAEA 122
>BU764568
Length = 420
Score = 34.3 bits (77), Expect = 0.013
Identities = 15/36 (41%), Positives = 21/36 (57%)
Frame = +3
Query: 25 TSGSCQFLEQALTGWSCRKQNTIALSTTEAEYESAA 60
TSG C + L W +KQ+ +A S+ EAEY + A
Sbjct: 165 TSGYCVLIGGNLISWKSKKQSVVAKSSAEAEYRAMA 272
>CO983062
Length = 390
Score = 33.5 bits (75), Expect = 0.023
Identities = 17/43 (39%), Positives = 26/43 (59%)
Frame = +3
Query: 15 YAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEYE 57
+A + +R+STS + F + L W +KQ A S+TEAEY+
Sbjct: 228 WASNIDDRRSTSRAAIFPGRNLISWWSKKQKVTARSSTEAEYQ 356
>TC221132 weakly similar to UP|O23529 (O23529) RETROTRANSPOSON like protein,
partial (5%)
Length = 799
Score = 33.1 bits (74), Expect = 0.030
Identities = 14/45 (31%), Positives = 26/45 (57%)
Frame = +1
Query: 12 DADYAGDKIERKSTSGSCQFLEQALTGWSCRKQNTIALSTTEAEY 56
DA++ + + +ST + ++ WSC+KQ+ I S+T+ EY
Sbjct: 265 DANWVDNTSDIRSTGAYVVYFGLSVISWSCKKQSIIDKSSTKVEY 399
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.332 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,527,659
Number of Sequences: 63676
Number of extensions: 24465
Number of successful extensions: 219
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 219
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 219
length of query: 61
length of database: 12,639,632
effective HSP length: 37
effective length of query: 24
effective length of database: 10,283,620
effective search space: 246806880
effective search space used: 246806880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146711.3


