

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146711.13 - phase: 0 /pseudo
(895 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
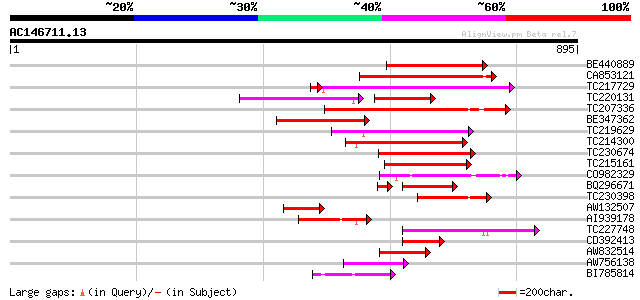
Score E
Sequences producing significant alignments: (bits) Value
BE440889 280 3e-75
CA853121 253 3e-67
TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 736... 218 3e-58
TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-li... 144 5e-54
TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, par... 189 4e-48
BE347362 179 6e-45
TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 171 2e-42
TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 169 6e-42
TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, p... 145 1e-34
TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%) 134 1e-31
CO982329 115 9e-26
BQ296671 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidop... 107 1e-25
TC230398 homologue to UP|O24147 (O24147) Kinesin-like protein, p... 111 2e-24
AW132507 102 7e-22
AI939178 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidop... 89 7e-18
TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partia... 80 4e-15
CD392413 similar to GP|13536981|dbj BY-2 kinesin-like protein 5 ... 74 2e-13
AW832514 72 1e-12
AW756138 similar to GP|6056195|gb|A kinesin-like protein {Arabid... 69 7e-12
BI785814 similar to GP|11761076|db kinesin-like protein {Oryza s... 67 3e-11
>BE440889
Length = 484
Score = 280 bits (715), Expect = 3e-75
Identities = 143/159 (89%), Positives = 149/159 (92%)
Frame = +2
Query: 595 HSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLS 654
H SR C+LCI VK KNL+NGE TKSKLWLVDL+GSERLAKTDVQGERLKEAQNINRSLS
Sbjct: 8 HQSRFLCLLCIAVKAKNLLNGESTKSKLWLVDLAGSERLAKTDVQGERLKEAQNINRSLS 187
Query: 655 ALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFA 714
ALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFA
Sbjct: 188 ALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFA 367
Query: 715 TRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEE 753
TRVRGVEL PVKKQIDT E+QK KAML+KARSECR K+E
Sbjct: 368 TRVRGVELGPVKKQIDTSEVQKMKAMLEKARSECRIKDE 484
>CA853121
Length = 677
Score = 253 bits (645), Expect = 3e-67
Identities = 129/217 (59%), Positives = 167/217 (76%)
Frame = +3
Query: 552 QNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKN 611
Q +G VPG++EA+V DVW L++G+ AR+VGS + NE SSRSHC+L + V +N
Sbjct: 12 QAVDGTQEVPGLIEARVYGTVDVWEKLKSGNQARSVGSTSANELSSRSHCLLRVTVLGEN 191
Query: 612 LMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIP 671
L+NG+ T+S LWLVDL+GSER+ KT+ +GERLKE+Q IN+SLSALGDVISALA+KS+HIP
Sbjct: 192 LINGQKTRSHLWLVDLAGSERVGKTEAEGERLKESQFINKSLSALGDVISALASKSAHIP 371
Query: 672 YRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDT 731
YRNSKLTH+LQ SLGGD KTLMFVQISP D+ ETL SLNFATRVRG+E P +KQ D
Sbjct: 372 YRNSKLTHILQSSLGGDCKTLMFVQISPGAADLTETLCSLNFATRVRGIESGPARKQTDL 551
Query: 732 GELQKTKAMLDKARSECRCKEESLRKLEESLQNIESK 768
EL K K M++K + + E+ RKL+++LQ ++ +
Sbjct: 552 TELNKYKQMVEKVKHD----EKETRKLQDNLQAMQMR 650
>TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 73641-79546,
partial (26%)
Length = 1075
Score = 218 bits (554), Expect(2) = 3e-58
Identities = 123/315 (39%), Positives = 186/315 (59%), Gaps = 5/315 (1%)
Frame = +1
Query: 487 KTFTMEG----TEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATG 542
KT+TM G ++ + GVNYR L LF +S+ R + Y++ V ++E+YNEQ+RDLL++
Sbjct: 37 KTYTMSGPGLSSKSDWGVNYRALHDLFHISQSRRSSIVYEVGVQMVEIYNEQVRDLLSSN 216
Query: 543 PASKRLEIKQNYEGHH-HVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHC 601
KRL I + + VP V++++DV ++ G RA + +NE SSRSH
Sbjct: 217 GPQKRLGIWNTAQPNGLAVPDASMHSVNSMADVLELMNIGLMNRATSATALNERSSRSHS 396
Query: 602 MLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVIS 661
+L + V+ +L + L LVDL+GSER+ +++ G+ LKEAQ+IN+SLSALGDVI
Sbjct: 397 VLSVHVRGTDLKTNTLLRGCLHLVDLAGSERVDRSEATGDMLKEAQHINKSLSALGDVIF 576
Query: 662 ALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVE 721
AL+ KSSH+PYRNSKLT LLQ SLGG +KTLMFVQ++P ET+S+L FA RV GVE
Sbjct: 577 ALSQKSSHVPYRNSKLTQLLQSSLGGQAKTLMFVQLNPDVASYSETVSTLKFAERVSGVE 756
Query: 722 LDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLEESLQNIESKAKGKDNIHKNLQE 781
L + + ++++ L + K+E + +L+ N G+ ++
Sbjct: 757 LGAARSNKEGRDVRELMEQLASLKDVIARKDEEIERLQSLKANHNGAKLGRISVRHGSSS 936
Query: 782 KIKELEGQIKLKTSM 796
+ G ++ T +
Sbjct: 937 PRRHSIGTPRISTRL 981
Score = 27.3 bits (59), Expect(2) = 3e-58
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +3
Query: 476 CIFAYGQTGTGKTFTMEGT 494
CIFAYGQTG+ K E T
Sbjct: 3 CIFAYGQTGSXKDIYDEWT 59
>TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-like protein
B), partial (37%)
Length = 1044
Score = 144 bits (363), Expect(2) = 5e-54
Identities = 85/207 (41%), Positives = 121/207 (58%), Gaps = 12/207 (5%)
Frame = +3
Query: 364 VQELVKQCEDLKVKYYEEMTQRKKLFNEVQEAKGNIRVFCRCRPLNKVEMSSGCTTVVDF 423
V EL ++ D + K E RKKL N + E KGNIRVFCR RPL E S + +
Sbjct: 75 VNELQRRLADAEYKLIEGERLRKKLHNTILELKGNIRVFCRVRPLLADESCSTEGRIFSY 254
Query: 424 DAAKD--GCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYG 481
+ + G LA K F FD+V+TP+ Q +VF + S +V S LDGY VCIFAYG
Sbjct: 255 PTSMETSGRAIDLAQNGQKHAFTFDKVFTPEASQEEVFVEISQLVQSALDGYKVCIFAYG 434
Query: 482 QTGTGKTFTM---EGTEQNRGVNYRTLEHLFRVSK-ERSETFSYDISVSVLEVYNEQIRD 537
QTG+GKT+TM G + +G+ R+LE +F+ + ++ + + Y++ VS+LE+YNE IRD
Sbjct: 435 QTGSGKTYTMMGRPGHPEEKGLIPRSLEQIFQTKQSQQPQGWKYEMQVSMLEIYNETIRD 614
Query: 538 LLAT------GPASKRLEIKQNYEGHH 558
L++T G K+ IK + G +
Sbjct: 615 LISTTTRMENGTPGKQYTIKHDTNGKY 695
Score = 86.7 bits (213), Expect(2) = 5e-54
Identities = 44/96 (45%), Positives = 63/96 (64%)
Frame = +1
Query: 577 VLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKT 636
+L +N+R+VG +NE SSRSH + + + N + + L L+DL+GSERL+K+
Sbjct: 757 LLNQAANSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQGVLNLIDLAGSERLSKS 936
Query: 637 DVQGERLKEAQNINRSLSALGDVISALAAKSSHIPY 672
G+RLKE Q IN+SLS+L DVI ALA K H+P+
Sbjct: 937 GSTGDRLKETQAINKSLSSLSDVIFALAKKEDHVPF 1044
>TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, partial (22%)
Length = 1055
Score = 189 bits (481), Expect = 4e-48
Identities = 107/296 (36%), Positives = 181/296 (61%), Gaps = 2/296 (0%)
Frame = +3
Query: 497 NRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLLATGPASK-RLEIKQNYE 555
N G+ R + LFR+ + + +S+ + ++E+Y + + DLL +L+IK++
Sbjct: 3 NPGLTPRAIAELFRILRRDNNKYSFSLKAYMVELYQDTLIDLLLPKNGKPLKLDIKKDST 182
Query: 556 GHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNG 615
G V V + I ++ +++Q GS R + +N+ SSRSH +L I++++ NL +
Sbjct: 183 GMVVVENVTVMSISTIEELNSIIQRGSERRHISGTQMNDESSRSHLILSIVIESTNLQSQ 362
Query: 616 ECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNS 675
K KL VDL+GSER+ K+ G +LKEAQ+IN+SLSALGDVIS+L++ H PYRN
Sbjct: 363 SVAKGKLSFVDLAGSERVKKSGSTGSQLKEAQSINKSLSALGDVISSLSSGGQHTPYRNH 542
Query: 676 KLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQ 735
KLT L+ DSLGG++KTLMFV ++P++ ++ ET +SL +A+RVR + DP K + + E+
Sbjct: 543 KLTMLMSDSLGGNAKTLMFVNVAPTESNLDETNNSLMYASRVRSIVNDP-NKNVSSKEVA 719
Query: 736 KTKAMLDKARSECRCKEESLRKLE-ESLQNIESKAKGKDNIHKNLQEKIKELEGQI 790
+ K ++ K+++ R LE + L+ I+ + K+N + N ++ ++ +I
Sbjct: 720 RLKKLV------AYWKQQAGRTLEYDDLEEIQDERPTKENGNGNR*KRSSQIATKI 869
>BE347362
Length = 448
Score = 179 bits (453), Expect = 6e-45
Identities = 91/150 (60%), Positives = 116/150 (76%), Gaps = 3/150 (2%)
Frame = +1
Query: 422 DFDAAKDGCLGILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYG 481
+F+++ D L ++ SSKK F+FD V+ P+D+Q VF +V SVLDGYNVCIFAYG
Sbjct: 1 NFESSSDNELQVICADSSKKQFKFDHVFGPEDNQETVFQQTKPIVTSVLDGYNVCIFAYG 180
Query: 482 QTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRDLL-- 539
QTGTGKTFTMEGT ++RGVNYRTLE LFR+++ER T Y++SVS+LEVYNE+IRDLL
Sbjct: 181 QTGTGKTFTMEGTPEHRGVNYRTLEELFRITEERHGTMKYELSVSMLEVYNEKIRDLLVE 360
Query: 540 -ATGPASKRLEIKQNYEGHHHVPGVVEAKV 568
+T P +K+LEIKQ EG VPG+VEA+V
Sbjct: 361 NSTQP-TKKLEIKQAAEGTQEVPGLVEARV 447
>TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (26%)
Length = 923
Score = 171 bits (432), Expect = 2e-42
Identities = 95/233 (40%), Positives = 139/233 (58%), Gaps = 9/233 (3%)
Frame = +1
Query: 508 LFRVSKE-RSETFSYDISVSVLEVYNEQIRDLLATGPASKRLEIKQNYEGH--------H 558
+F+ S+ + + + Y + VS+ E+YNE IRDLL++ +S +
Sbjct: 10 IFQTSQSLKDQGWKYTMHVSIYEIYNETIRDLLSSNRSSGNDHTRTENSAPTPSKQHTIK 189
Query: 559 HVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECT 618
H + +V + ++ ++LQ + +R+VG +NE SSRSH + + + +N +
Sbjct: 190 HESDLATLEVCSAEEISSLLQQAAQSRSVGRTQMNEQSSRSHFVFKLRISGRNEKTEKQV 369
Query: 619 KSKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLT 678
+ L L+DL+GSERL+++ G+RLKE Q IN+SLS+L DVI ALA K H+P+RNSKLT
Sbjct: 370 QGVLNLIDLAGSERLSRSGATGDRLKETQAINKSLSSLSDVIFALAKKEEHVPFRNSKLT 549
Query: 679 HLLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELDPVKKQIDT 731
H LQ LGGDSKTLMFV ISP GE+L SL FA RV E+ ++Q T
Sbjct: 550 HFLQPYLGGDSKTLMFVNISPDQSSAGESLCSLRFAARVNACEIGIPRRQTQT 708
>TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (27%)
Length = 903
Score = 169 bits (427), Expect = 6e-42
Identities = 95/207 (45%), Positives = 128/207 (60%), Gaps = 14/207 (6%)
Frame = +2
Query: 530 VYNEQIRDLLATGPAS--------------KRLEIKQNYEGHHHVPGVVEAKVDNISDVW 575
+YNE IRDLLAT +S K+ IK + G+ HV + V ++ +V
Sbjct: 2 IYNETIRDLLATNKSSADGTPTRVENGTPGKQYMIKHDANGNTHVSDLTVVDVQSVKEVA 181
Query: 576 TVLQAGSNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAK 635
+L +++R+VG +NE SSRSH + + + N + + L L+DL+GSERL++
Sbjct: 182 FLLNQAASSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQGILNLIDLAGSERLSR 361
Query: 636 TDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFV 695
+ G+RLKE Q IN+SLS+L DVI ALA K HIP+RNSKLT+LLQ LGGDSKTLMFV
Sbjct: 362 SGSTGDRLKETQAINKSLSSLSDVIFALAKKEDHIPFRNSKLTYLLQPCLGGDSKTLMFV 541
Query: 696 QISPSDQDVGETLSSLNFATRVRGVEL 722
ISP GE+L SL FA+RV E+
Sbjct: 542 NISPDQASSGESLCSLRFASRVNACEI 622
>TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, partial (20%)
Length = 811
Score = 145 bits (365), Expect = 1e-34
Identities = 77/153 (50%), Positives = 101/153 (65%)
Frame = +1
Query: 582 SNARAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGE 641
S +R+VG ++NE SSRSH + + + N + + L L+DL+GSERL+++ G+
Sbjct: 85 SCSRSVGRTHMNEQSSRSHFVXTLRISGTNSNTDQQVQGVLNLIDLAGSERLSRSGATGD 264
Query: 642 RLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSD 701
RLKE Q IN+SLS+L DVI ALA K H+P+RNSKLT+LLQ LGGDSKTLMFV ISP
Sbjct: 265 RLKETQAINKSLSSLSDVIFALAKKQEHVPFRNSKLTYLLQPCLGGDSKTLMFVNISPDP 444
Query: 702 QDVGETLSSLNFATRVRGVELDPVKKQIDTGEL 734
GE+L SL FA V E+ ++Q T L
Sbjct: 445 SSTGESLCSLRFAAGVNACEIGIPRRQTSTRSL 543
>TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%)
Length = 692
Score = 134 bits (338), Expect = 1e-31
Identities = 72/137 (52%), Positives = 92/137 (66%)
Frame = +2
Query: 592 VNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSGSERLAKTDVQGERLKEAQNINR 651
+NE SSRSH + + + N + L L+DL+GSERL+K+ G+RLKE Q IN+
Sbjct: 41 MNEQSSRSHFVFTLRIYGVNESTDXXVQGVLNLIDLAGSERLSKSGSTGDRLKETQAINK 220
Query: 652 SLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQDVGETLSSL 711
SLS+L DVI ALA K H+P+RNSKLT+LLQ LGG S TLMFV ISP VGE+L SL
Sbjct: 221 SLSSLSDVIFALAKKEDHVPFRNSKLTYLLQPCLGGYSNTLMFVNISPDPSSVGESLCSL 400
Query: 712 NFATRVRGVELDPVKKQ 728
FA+RV E+ ++Q
Sbjct: 401 RFASRVNACEIGTPRRQ 451
>CO982329
Length = 852
Score = 115 bits (288), Expect = 9e-26
Identities = 76/228 (33%), Positives = 131/228 (57%), Gaps = 5/228 (2%)
Frame = -3
Query: 585 RAVGSNNVNEHSSRSHCMLCIMVKT----KNLMNGECTKSKLWLVDLSGSERLAKTDVQG 640
R VGS N N SSRSH + + +++ +N T S+L L+DL+GSE ++ + G
Sbjct: 832 RHVGSTNFNLLSSRSHTIFSLTIESSPCGENNEGEAITLSQLNLIDLAGSES-SRAETTG 656
Query: 641 ERLKEAQNINRSLSALGDVISALA-AKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISP 699
R +E IN+SL LG VIS L K+SHIPYR+SKLT LLQ SL G + + ++P
Sbjct: 655 MRRREGSYINKSLLTLGTVISKLTEGKASHIPYRDSKLTRLLQSSLSGHGRISLICTVTP 476
Query: 700 SDQDVGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLDKARSECRCKEESLRKLE 759
S + ET ++L FA R + +E+ + I K+++ K + E + +E L ++
Sbjct: 475 SSSNAEETHNTLKFAHRAKHIEIQAAQNTI-----IDEKSLIKKYQHEIQFLKEELEQMR 311
Query: 760 ESLQNIESKAKGKDNIHKNLQEKIKELEGQIKLKTSMQNQSEKQVSQL 807
+ + +++SK + + L++K++ +GQ+KL++ ++ + E + + L
Sbjct: 310 QGIVSVQSKETVEVDF-VLLKQKLE--DGQVKLQSRLEEEEEAKAALL 176
>BQ296671 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidopsis
thaliana}, partial (20%)
Length = 422
Score = 107 bits (266), Expect(2) = 1e-25
Identities = 54/87 (62%), Positives = 65/87 (74%)
Frame = +3
Query: 620 SKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTH 679
SKLW++DL GSERL KT +G L E + IN SLSAL DV++AL K H+PYRNSKLT
Sbjct: 162 SKLWMIDLGGSERLLKTGAKGLTLDEGRAINLSLSALADVVAALKRKRCHVPYRNSKLTQ 341
Query: 680 LLQDSLGGDSKTLMFVQISPSDQDVGE 706
+L+DSLG SK LM V ISPS++DV E
Sbjct: 342 ILKDSLGYGSKVLMLVHISPSEEDVCE 422
Score = 28.5 bits (62), Expect(2) = 1e-25
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = +1
Query: 581 GSNARAVGSNNVNEHSSRSHCML 603
G R+ NVNE SSRSHC +
Sbjct: 28 GKRFRSTSWTNVNEASSRSHCFI 96
>TC230398 homologue to UP|O24147 (O24147) Kinesin-like protein, partial (10%)
Length = 599
Score = 111 bits (277), Expect = 2e-24
Identities = 59/119 (49%), Positives = 86/119 (71%), Gaps = 2/119 (1%)
Frame = +3
Query: 644 KEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHLLQDSLGGDSKTLMFVQISPSDQD 703
KEAQ+IN+SLSALGDVISAL++ HIPYRN KLT L+ DSLGG++KTLMFV +SP +
Sbjct: 3 KEAQSINKSLSALGDVISALSSGGQHIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPVESS 182
Query: 704 VGETLSSLNFATRVRGVELDPVKKQIDTGELQKTKAMLD--KARSECRCKEESLRKLEE 760
+ ET +SL +A+RVR + DP K + + E+ + K ++ K ++ R ++E L +++E
Sbjct: 183 LDETHNSLMYASRVRSIVNDP-SKNVSSKEIARLKKLIGYWKEQAGRRGEDEDLEEIQE 356
>AW132507
Length = 196
Score = 102 bits (254), Expect = 7e-22
Identities = 48/65 (73%), Positives = 57/65 (86%)
Frame = +2
Query: 433 ILATGSSKKLFRFDRVYTPKDDQVDVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTME 492
I +GS+KK F FDRVYTPKDDQ+D+ + SS+VISVL+GYNVCIF+YGQTGTGKTFTME
Sbjct: 2 IKTSGSTKKSFIFDRVYTPKDDQLDLCSYTSSIVISVLNGYNVCIFSYGQTGTGKTFTME 181
Query: 493 GTEQN 497
GT+ N
Sbjct: 182 GTQHN 196
>AI939178 similar to GP|18087642|gb AT5g27950/F15F15_20 {Arabidopsis
thaliana}, partial (19%)
Length = 406
Score = 89.4 bits (220), Expect = 7e-18
Identities = 49/124 (39%), Positives = 77/124 (61%), Gaps = 10/124 (8%)
Frame = +3
Query: 457 DVFADASSMVISVLDGYNVCIFAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFR-VSKER 515
+VF + ++ S +DG+NVC+FAYGQTGTGKTFTM+GT Q G+ R LE LFR S +
Sbjct: 3 EVFVEVEPILRSAMDGHNVCVFAYGQTGTGKTFTMDGTNQEPGIIPRALEELFRQASLDN 182
Query: 516 SETFSYDISVSVLEVYNEQIRDLLATGPASK---------RLEIKQNYEGHHHVPGVVEA 566
S +F++ ++S+LEVY +RDLL+ + + L I+ + +G + G+ E
Sbjct: 183 SSSFTF--TMSMLEVYMGNLRDLLSPRQSGRPHEQYMTKCNLNIQTDPKGLIEIEGLSEV 356
Query: 567 KVDN 570
++ +
Sbjct: 357 QISD 368
>TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partial (34%)
Length = 1192
Score = 80.1 bits (196), Expect = 4e-15
Identities = 69/236 (29%), Positives = 114/236 (48%), Gaps = 20/236 (8%)
Frame = +3
Query: 621 KLWLVDLSGSERLAKTDVQGERLK-EAQNINRSLSALGDVISALAAKSSHIPYRNSKLTH 679
KL +DL+GSER A T ++ + E IN+SL AL + I AL HIP+R SKLT
Sbjct: 3 KLSFIDLAGSERGADTTDNDKQTRIEGAEINKSLLALKECIRALDNDQGHIPFRGSKLTE 182
Query: 680 LLQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVEL-DPVKKQIDTGELQKTK 738
+L+DS G+S+T+M ISPS TL++L +A RV+ + + KK + + +
Sbjct: 183 VLRDSFVGNSRTVMISCISPSTGSCEHTLNTLRYADRVKSLSKGNNSKKDVLSSNFNLKE 362
Query: 739 AMLDKARS------ECRCKE--------ESLRKLEESLQNIES--KAKGKDNIHKNLQEK 782
+ S E R + + EE + ++ K GK + +K
Sbjct: 363 SSTVPLSSVTGSAYEDRVTDGWPDENEGDDFSPSEEYYEQVKPPLKKNGKMESYATTDDK 542
Query: 783 IKELEGQIKLK--TSMQNQSEKQVSQLCERLKGKEETCCTLQHKVKELERKIKEQL 836
+K+ GQIK K ++ Q+ L L+ +E+ + +V+E ++E++
Sbjct: 543 LKKPSGQIKWKDLPKVEPQTTHAEDDLNALLQEEEDLVNAHRTQVEETMNIVREEM 710
>CD392413 similar to GP|13536981|dbj BY-2 kinesin-like protein 5 {Nicotiana
tabacum}, partial (13%)
Length = 394
Score = 74.3 bits (181), Expect = 2e-13
Identities = 37/67 (55%), Positives = 49/67 (72%)
Frame = -3
Query: 620 SKLWLVDLSGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTH 679
SKLW++DL GS++L KT + L E + IN SL ALGDV++AL K H+P+RNS LT
Sbjct: 296 SKLWMIDLEGSKQLLKTGAKELTLDEGRAINLSLLALGDVVAALKRKRCHVPFRNS*LTL 117
Query: 680 LLQDSLG 686
+L+DSLG
Sbjct: 116 ILKDSLG 96
>AW832514
Length = 412
Score = 71.6 bits (174), Expect = 1e-12
Identities = 39/80 (48%), Positives = 56/80 (69%), Gaps = 1/80 (1%)
Frame = +3
Query: 585 RAVGSNNVNEHSSRSHCMLCIMVKTKNLMNGECTK-SKLWLVDLSGSERLAKTDVQGERL 643
RAVG +N SSRSHC+ ++ + L + T+ KL LVDL+GSE++ KT +G L
Sbjct: 156 RAVGETQMNVASSRSHCIYIFTIQQEFLSRDKRTRFGKLILVDLAGSEKVEKTGAEGRVL 335
Query: 644 KEAQNINRSLSALGDVISAL 663
+EA+ IN+SLSALG+VI++L
Sbjct: 336 EEAKTINKSLSALGNVINSL 395
>AW756138 similar to GP|6056195|gb|A kinesin-like protein {Arabidopsis
thaliana}, partial (8%)
Length = 313
Score = 69.3 bits (168), Expect = 7e-12
Identities = 40/102 (39%), Positives = 60/102 (58%)
Frame = +1
Query: 528 LEVYNEQIRDLLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAV 587
+E+YNEQ+RDLLA +LEI+ + +P + V + +DV T+++ G RAV
Sbjct: 10 VEIYNEQVRDLLAEDKTDNKLEIRSCNDDGLSLPDAILHSVKSPTDVMTLIKLGEVNRAV 189
Query: 588 GSNNVNEHSSRSHCMLCIMVKTKNLMNGECTKSKLWLVDLSG 629
S +N SSRSH +L + V K+ +G +S L LVDL+G
Sbjct: 190 SSTAMNNRSSRSHSVLTVHVNGKD-TSGSSIRSCLHLVDLAG 312
>BI785814 similar to GP|11761076|db kinesin-like protein {Oryza sativa
(japonica cultivar-group)}, partial (19%)
Length = 421
Score = 67.0 bits (162), Expect = 3e-11
Identities = 37/131 (28%), Positives = 72/131 (54%)
Frame = +3
Query: 478 FAYGQTGTGKTFTMEGTEQNRGVNYRTLEHLFRVSKERSETFSYDISVSVLEVYNEQIRD 537
FAYGQTG+GKT+TM + + + + R+ + + VS E+Y ++ D
Sbjct: 6 FAYGQTGSGKTYTM------KPLPLKASRDILRLMHHTYRNQGFQLFVSFFEIYGGKLFD 167
Query: 538 LLATGPASKRLEIKQNYEGHHHVPGVVEAKVDNISDVWTVLQAGSNARAVGSNNVNEHSS 597
LL K+L ++++ + + G+ E +V ++ ++ +++ G++ R+ G+ NE SS
Sbjct: 168 LLND---RKKLCMREDGKQQVCIVGLQEYRVSDVENIKDLIEKGNSTRSTGTTGANEESS 338
Query: 598 RSHCMLCIMVK 608
RSH +L + +K
Sbjct: 339 RSHAILQLAIK 371
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,911,596
Number of Sequences: 63676
Number of extensions: 391020
Number of successful extensions: 2210
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 2174
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2194
length of query: 895
length of database: 12,639,632
effective HSP length: 106
effective length of query: 789
effective length of database: 5,889,976
effective search space: 4647191064
effective search space used: 4647191064
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146711.13


