

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.5 + phase: 0 /pseudo
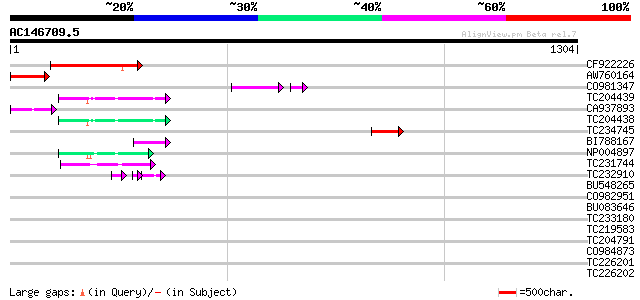
(1304 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922226 191 2e-48
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 99 1e-20
CO981347 92 1e-20
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 61 3e-09
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 60 8e-09
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 59 1e-08
TC234745 weakly similar to UP|Q9FFM0 (Q9FFM0) Copia-like retrotr... 55 2e-07
BI788167 51 4e-06
NP004897 gag-protease polyprotein 50 5e-06
TC231744 47 7e-05
TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial ... 32 2e-04
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 43 0.001
CO982951 42 0.001
BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 42 0.002
TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, parti... 41 0.004
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 38 0.026
TC204791 weakly similar to UP|O82196 (O82196) Copia-like retroel... 35 0.17
CO984873 35 0.22
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 34 0.37
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 34 0.37
>CF922226
Length = 667
Score = 191 bits (485), Expect = 2e-48
Identities = 99/219 (45%), Positives = 145/219 (66%), Gaps = 6/219 (2%)
Frame = -3
Query: 93 MTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKVLHLLCAL 152
MTKSL +R KQ LY ++M E + + EQL FNK+I DL NIDV ++DED+ L LLC L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 153 PRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNK-G 211
P+S+ +FK+T+L+G++ +++L+EVQ AL +KEL + KE K SGEGL +RG++ K
Sbjct: 485 PKSYSHFKETLLFGRD-SVSLDEVQTALNSKELNERKEKKSSASGEGLT-ARGKTFKKDS 312
Query: 212 KGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGG-----ESSVQIASK 266
+ K + +++ G+GN + +C+ C GH +K CPER+ NGG +S +
Sbjct: 311 EFDKKKQKPENQKNGEGNIFKIRCYHCKKEGHTRKVCPERQKNGGSNNRKKDSGNAAIVQ 132
Query: 267 DEGYESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFE 305
D+GYESA AL V+ PE W++DSGCS+H+ P K +FE
Sbjct: 131 DDGYESAEALMVSEKNPETKWIMDSGCSWHMTPNKSWFE 15
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 99.0 bits (245), Expect = 1e-20
Identities = 51/91 (56%), Positives = 68/91 (74%), Gaps = 1/91 (1%)
Frame = +3
Query: 2 MGS-KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDKAV 60
MGS K+++EKFTG NDFGL +KMRA+L+QQ VEAL GE ++ + +K + KA
Sbjct: 75 MGSAKYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAY 254
Query: 61 SAIILCLGDKVLREVSREATAVSMWNKLDSL 91
+AIIL LGDKVLR+VS+E TAV +W+KL+ L
Sbjct: 255 NAIILSLGDKVLRQVSKETTAVGVWSKLEVL 347
>CO981347
Length = 624
Score = 92.4 bits (228), Expect(2) = 1e-20
Identities = 60/120 (50%), Positives = 70/120 (58%)
Frame = +3
Query: 511 RRKVMLLKNSRNGIHL*KIRLVPN*KC*ELTMAWSLFQNSLMSFAGRKV*RGIEPWHTHL 570
+ K KNS N I L +I LV N*K * LTMAWSLF +S MSFAG+ +G + TH
Sbjct: 90 KNKSESFKNSENDILLLEINLVQN*KF*GLTMAWSLF*SSSMSFAGK*ASKGTK*SLTHH 269
Query: 571 NRMVLLKE*TGLCWSA*GVCC*KLDCPRVSGERLLVLQHI*LTDVHQQG*ISRHLWRFGV 630
+RMV K * W *G C * DC R GE+L HI*L D Q *+SRH W+ GV
Sbjct: 270 SRMV*QKG*I*PFWKE*GACY*VQDCQRPFGEKLQTQHHI*LIDALHQP*VSRHQWKLGV 449
Score = 26.9 bits (58), Expect(2) = 1e-20
Identities = 18/39 (46%), Positives = 20/39 (51%)
Frame = +1
Query: 647 LPMSGKTNLMLEL*SVFSWVILKV*KVIDCGRWNLENQN 685
L M K + M L*SV S ILK K G WNL Q+
Sbjct: 499 LIMLNKESWMQGL*SVCSLAILKELKGTSYGNWNLVRQD 615
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 61.2 bits (147), Expect = 3e-09
Identities = 72/276 (26%), Positives = 111/276 (40%), Gaps = 20/276 (7%)
Frame = +1
Query: 113 VESKPIMEQLTEFNKIIDDL-ANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTI 171
++S+ I++Q + K+I DL A + + E+ ++ + L EN ++ +G+
Sbjct: 1135 IKSEKILQQEAQLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1314
Query: 172 TLEEV-----------------QAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGK 214
TL+EV ++A RT +T+F K + G +S+ RS + G +
Sbjct: 1315 TLDEVLLLGKNAGNQRGLGFNPKSAGRTT-MTEFVPAK---NRTGATMSQHRSRHHGMQQ 1482
Query: 215 GKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAG 274
K+ R K R G K F H GH P + S
Sbjct: 1483 KKSKRKKWRCHYCGKYGHIKPFCYHLHGH-----PHHGTQSSNSRKKMMWVPKHKAVSLV 1647
Query: 275 ALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLK 334
T ++ W LDSGCS H+ KE+ +E V G+ KI GMG K
Sbjct: 1648 VHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMG----K 1815
Query: 335 MFDDRDFLLKNVRYIPELKRNLISISMF--DGLELN 368
+ D L V + L NLISIS +G +N
Sbjct: 1816 LVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVN 1923
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 59.7 bits (143), Expect = 8e-09
Identities = 33/106 (31%), Positives = 59/106 (55%)
Frame = -2
Query: 3 GSKWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDKAVSA 62
G+K+++ KF G+ +F LW+ +++ +L Q ++ L+ + E E+ ++ +
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSKSNNTEALDWE--EL*ERTATT 142
Query: 63 IILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLY 108
I LCL D+ L + A +W KL+S YM KSL ++ L Q+LY
Sbjct: 141 IRLCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 58.9 bits (141), Expect = 1e-08
Identities = 70/276 (25%), Positives = 110/276 (39%), Gaps = 20/276 (7%)
Frame = +1
Query: 113 VESKPIMEQLTEFNKIIDDL-ANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTI 171
++S+ I++Q + K+I +L A + + E+ ++ + L EN ++ +G+
Sbjct: 1138 IKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1317
Query: 172 TLEEV-----------------QAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGK 214
L+EV ++A RT +T+F K + G +S+ RS + G +
Sbjct: 1318 MLDEVLQLGKNVGNQRGLGFNHKSAGRTT-MTEFVPAK---NSTGATMSQHRSRHHGTQQ 1485
Query: 215 GKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAG 274
K+ R K R G K F H GH P + S
Sbjct: 1486 KKSKRKKWRCHYCGKYGHIKPFCYHLHGH-----PHHGTQSSSSGRKMMWVPKHKIVSLV 1650
Query: 275 ALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLK 334
T ++ W LDSGCS H+ KE+ +E V G+ KI GMG K
Sbjct: 1651 VHTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMG----K 1818
Query: 335 MFDDRDFLLKNVRYIPELKRNLISISMF--DGLELN 368
+ D L V + L NLISIS +G +N
Sbjct: 1819 LVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVN 1926
Score = 30.8 bits (68), Expect = 4.1
Identities = 31/105 (29%), Positives = 44/105 (41%)
Frame = +2
Query: 540 LTMAWSLFQNSLMSFAGRKV*RGIEPWHTHLNRMVLLKE*TGLCWSA*GVCC*KLDCPRV 599
+TMA SL SL++ A K H N+M LK TGLC G C + P +
Sbjct: 2462 VTMAESLKTASLLNSAHLKASLMSSLQPLHHNKMA*LKGKTGLCKKLLGSCFMPKNFPII 2641
Query: 600 SGERLLVLQHI*LTDVHQQG*ISRHLWRFGVEDRQTTLT*KSSEL 644
SG + T+ H + + H + G Q + T S E+
Sbjct: 2642 SGLKP*TQHATSTTESHLEEGLQPHCMKSGKGGSQLSSTSTSLEV 2776
>TC234745 weakly similar to UP|Q9FFM0 (Q9FFM0) Copia-like retrotransposable
element, partial (5%)
Length = 494
Score = 55.1 bits (131), Expect = 2e-07
Identities = 33/73 (45%), Positives = 45/73 (61%)
Frame = -1
Query: 833 EASGCSRRKKVYQGSKHQGIRQDL*RRVLLRWKGLITMKSFHR**NIVL*GYLWRLLTCI 892
+AS SR++K +Q + +G RQDL RVL +W LI MK H *NI+L*G W+L +
Sbjct: 260 DASRSSRKRKTFQV*RSRGTRQDLWLRVLHKWTELIIMKFSHLW*NIIL*GS*WQL*ISM 81
Query: 893 ILS*NKWM*RPHF 905
I S + W+ R F
Sbjct: 80 IWSYHNWLPRQLF 42
>BI788167
Length = 421
Score = 50.8 bits (120), Expect = 4e-06
Identities = 32/89 (35%), Positives = 50/89 (55%), Gaps = 3/89 (3%)
Frame = +2
Query: 284 EKSWVLDSGCS---YHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRD 340
+ S +LD G S +H+ P E F T E G V +GN+ A +I G+GTI+LKM+D
Sbjct: 29 DNS*LLDIGFSCATWHMTPC*E*FCTYETILEGFVFMGNDHALEIVGVGTIKLKMYDGIV 208
Query: 341 FLLKNVRYIPELKRNLISISMFDGLELNV 369
++ V ++ LK+N + + D LE +
Sbjct: 209 RTIQGVLHVKGLKKNQLFVGKLDDLECKI 295
>NP004897 gag-protease polyprotein
Length = 1923
Score = 50.4 bits (119), Expect = 5e-06
Identities = 56/236 (23%), Positives = 91/236 (37%), Gaps = 17/236 (7%)
Frame = +1
Query: 113 VESKPIMEQLTEFNKIIDDL-ANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTI 171
++S+ I++Q + K+I +L A + + E+ ++ + L EN ++ +G+
Sbjct: 1138 IKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKMLNKGSD 1317
Query: 172 TLEEV---------QAALRTKE-------LTKFKELKVDDSGEGLNISRGRSHNKGKGKG 215
L+EV Q L +T+F K+ G +S+ RS + G +
Sbjct: 1318 MLDEVLQLGKNVGNQRGLGFNHKSAGRITMTEFVPAKIST---GATMSQHRSRHHGTQQK 1488
Query: 216 KNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGA 275
K+ R K R G K F H GH P + S
Sbjct: 1489 KSKRKKWRCHYCGKYGHIKPFCYHLHGH-----PHHGTQSSSSRRKMMWVPKHKIVSLVV 1653
Query: 276 LTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTI 331
T ++ W LDSGCS H+ KE+ +E V G+ KI GMG +
Sbjct: 1654 HTSLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKL 1821
>TC231744
Length = 794
Score = 46.6 bits (109), Expect = 7e-05
Identities = 49/225 (21%), Positives = 92/225 (40%), Gaps = 7/225 (3%)
Frame = +2
Query: 118 IMEQLTEFNKIIDDLANIDVNLEDEDKVLHL-LCALPRSFENFKDTMLYGKEGTITLEEV 176
I E + E + + L ++ + L ED +HL L +LP F FK + K+ E +
Sbjct: 86 IREYIMEISNLASKLKSLKLEL-GEDLFVHLVLISLPAHFGQFKVSYNTQKDKWSLNELI 262
Query: 177 QAALRTKELTKFKELKVDDSGEGLNISRGRS---HNKGKGKGKNSRSKSR-SKGDGNKTQ 232
++ +E + R R+ HNK + K K+ K+ K +
Sbjct: 263 SHCVQEEE----------------RL*RDRTESAHNKKRKKTKDVAEKTS*QKKQQKDEE 394
Query: 233 YKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGALTVTSWEPEKSWVLDSG 292
+ C+ C H KK CP + + KD+ + ++ P+ +W +DSG
Sbjct: 395 FTCYFCKKSRHMKKKCP--------KYAAWRVKKDKFLTLVCSEVNLAFVPKDTWWVDSG 550
Query: 293 CSYHICPRKE--YFETLELKEGGVVRLGNNKACKIQGMGTIRLKM 335
+ HI + + L + + +G+ K ++ + T RL++
Sbjct: 551 ATTHISMTMQGCLWSRLPSDDERFIFMGDGKKVAVEAIETFRLQL 685
>TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial (3%)
Length = 690
Score = 32.3 bits (72), Expect(3) = 2e-04
Identities = 16/56 (28%), Positives = 31/56 (54%)
Frame = +2
Query: 303 YFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLIS 358
+ E + +G +V +GN I G+G + L +F L +V ++P +++NL+S
Sbjct: 206 FMEFRPIDDGSIVNMGNVATEPILGLGCVNL-VFTSGKSLYLDVLFVPGIRKNLLS 370
Score = 27.3 bits (59), Expect(3) = 2e-04
Identities = 14/36 (38%), Positives = 16/36 (43%), Gaps = 1/36 (2%)
Frame = +3
Query: 235 CFICHNPGHFKKDCPERK-DNGGGESSVQIASKDEG 269
C C PGH K+DC K N G S K +G
Sbjct: 33 CSKCGKPGHLKRDCRVFKGKNKAGPSGSNDPKKQQG 140
Score = 23.5 bits (49), Expect(3) = 2e-04
Identities = 7/23 (30%), Positives = 13/23 (56%)
Frame = +1
Query: 282 EPEKSWVLDSGCSYHICPRKEYF 304
+ + +W DSG + H+C + F
Sbjct: 139 DDDVAWWFDSGATSHVCKDRRLF 207
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 42.7 bits (99), Expect = 0.001
Identities = 24/90 (26%), Positives = 45/90 (49%), Gaps = 3/90 (3%)
Frame = -3
Query: 276 LTVTSWEPEKSWVLDSGCSYHICPRKEYFETL-ELKEGGVVRLGNNKACKI--QGMGTIR 332
LT ++ P +SW DSG S+H+ + + + +E + +GN + I G+ T
Sbjct: 650 LTSSAATPSQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLSTFS 471
Query: 333 LKMFDDRDFLLKNVRYIPELKRNLISISMF 362
+ +L N+ ++P + +NLI +S F
Sbjct: 470 SPINPQFSLVLSNLLFVPTITKNLIRVSQF 381
>CO982951
Length = 757
Score = 42.4 bits (98), Expect = 0.001
Identities = 21/48 (43%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = -3
Query: 206 RSHNKGKGKGK-NSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 252
+ H+KGKG K N+ SK K + C C GHF+KDCP+RK
Sbjct: 308 KKHDKGKGPLKINNNSKQIQKKTSKRNN--CHFCGKSGHFQKDCPKRK 171
>BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (19%)
Length = 428
Score = 42.0 bits (97), Expect = 0.002
Identities = 22/74 (29%), Positives = 40/74 (53%)
Frame = +1
Query: 286 SWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKN 345
SW++DSGC+ H+ +E F L+ V++ N ++G T+ + L+ N
Sbjct: 70 SWLIDSGCTNHMTYDRELFTELDEVVFSKVKIRNEAYIDVKGKETVAI*GHTGLK-LISN 246
Query: 346 VRYIPELKRNLISI 359
V Y+ E+ +NL+S+
Sbjct: 247 VLYVSEISQNLLSV 288
>TC233180 similar to UP|Q944K0 (Q944K0) At1g18030/T10F20_3, partial (11%)
Length = 916
Score = 40.8 bits (94), Expect = 0.004
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 1/90 (1%)
Frame = +2
Query: 271 ESAGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGV-VRLGNNKACKIQGMG 329
+S+ L+ + E ++DSG + H+ P YF + G + + N I G G
Sbjct: 404 KSSSFLSFNASGTENI*IIDSGVTDHMTPHSSYFSSYTFLIGNQHIIVANGSHIPIIGCG 583
Query: 330 TIRLKMFDDRDFLLKNVRYIPELKRNLISI 359
I+L+ L NV Y+P+L NL+SI
Sbjct: 584 NIQLQ----SSLHLNNVLYVPKLSNNLLSI 661
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 38.1 bits (87), Expect = 0.026
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 1/95 (1%)
Frame = +2
Query: 165 YGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRS 224
YG +G +V +A+R++ F RG +G+G G+ + R
Sbjct: 257 YGDDGRTMAVDVTSAVRSRLPGGF---------------RGGGGGRGRGGGRYGGGEGRG 391
Query: 225 KGDGNKTQY-KCFICHNPGHFKKDCPERKDNGGGE 258
+G G + +C+ C GH +DC + GGG+
Sbjct: 392 RGFGRRGGGPECYNCGRIGHLARDCYHGQGGGGGD 496
Score = 37.4 bits (85), Expect = 0.044
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +2
Query: 201 NISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCP 249
+++R H +G G G + R++ R G G CF C GHF ++CP
Sbjct: 449 HLARDCYHGQGGGGGDDGRNRRRGGGGGGGGG--CFNCGEEGHFARECP 589
>TC204791 weakly similar to UP|O82196 (O82196) Copia-like retroelement pol
polyprotein, partial (4%)
Length = 880
Score = 35.4 bits (80), Expect = 0.17
Identities = 24/55 (43%), Positives = 30/55 (53%)
Frame = +2
Query: 1245 IRCIMRGQSTLTFVCTSSET*LRLRRSW*RKWHRKTIQQTCSPNHYQDQDSSIAW 1299
I+ IMRG STLT TSS + + S RK R IQQ CS + + + S I W
Sbjct: 305 IKSIMRGLSTLTLDTTSS---ILRKGSKSRKLIRGRIQQICSRSRF*ETSSCIIW 460
>CO984873
Length = 754
Score = 35.0 bits (79), Expect = 0.22
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = -2
Query: 213 GKGKNSRSKSRSKGDGN---KTQYKCFICHNPGHFKKDCPERK 252
G K ++K++ K K + KCF C+ GH KKDC + K
Sbjct: 432 GMKKGDQAKNKGKITAKPVIKNESKCFFCNKKGHIKKDCSKFK 304
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 34.3 bits (77), Expect = 0.37
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Frame = +1
Query: 207 SHN-KGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNG 255
SHN +G G G+ RS + KC+ C PGHF ++C R +G
Sbjct: 184 SHNSRGGGGGRGGRSGG--------SDLKCYECGEPGHFARECRMRGGSG 309
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 34.3 bits (77), Expect = 0.37
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Frame = +3
Query: 207 SHN-KGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNG 255
SHN +G G G+ RS + KC+ C PGHF ++C R +G
Sbjct: 771 SHNSRGGGGGRGGRSGG--------SDLKCYECGEPGHFARECRMRGGSG 896
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.357 0.158 0.567
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,062,111
Number of Sequences: 63676
Number of extensions: 871631
Number of successful extensions: 9146
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 4960
Number of HSP's successfully gapped in prelim test: 314
Number of HSP's that attempted gapping in prelim test: 3911
Number of HSP's gapped (non-prelim): 5690
length of query: 1304
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1196
effective length of database: 5,762,624
effective search space: 6892098304
effective search space used: 6892098304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146709.5


