

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.1 - phase: 0 /pseudo
(495 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
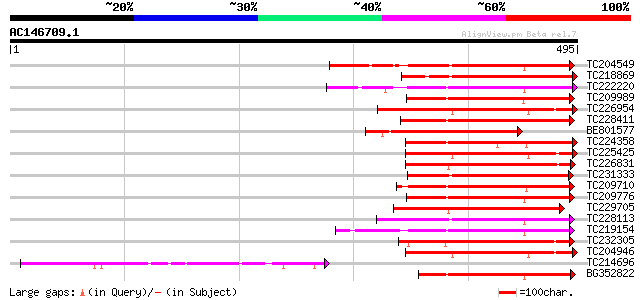
Score E
Sequences producing significant alignments: (bits) Value
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 153 2e-37
TC218869 similar to UP|Q40543 (Q40543) Protein-serine/threonine ... 149 4e-36
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 149 4e-36
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 147 1e-35
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 145 5e-35
TC228411 similar to UP|Q94JZ6 (Q94JZ6) Protein kinase-like prote... 144 7e-35
BE801577 similar to PIR|T49986|T499 lectin-like protein kinase-l... 143 2e-34
TC224358 similar to UP|Q8L7V7 (Q8L7V7) AT4g02010/T10M13_2, parti... 143 2e-34
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 143 2e-34
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 142 3e-34
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 142 4e-34
TC209710 similar to UP|Q9LPF9 (Q9LPF9) T3F20.25 protein, partial... 142 4e-34
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 140 2e-33
TC229705 similar to UP|Q9LFP7 (Q9LFP7) Serine/threonine specific... 139 2e-33
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 139 2e-33
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 139 3e-33
TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase... 139 4e-33
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 138 6e-33
TC214696 similar to UP|Q8L683 (Q8L683) Lectin precursor, partial... 138 6e-33
BG352822 similar to GP|20453162|g At5g56885 {Arabidopsis thalian... 137 1e-32
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 153 bits (386), Expect = 2e-37
Identities = 89/216 (41%), Positives = 138/216 (63%), Gaps = 2/216 (0%)
Frame = +3
Query: 280 TSGDSKTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDL 339
+SG S +V+ +TV+ + ++G+ L A +K++ S K+ + A + ++ D L
Sbjct: 618 SSGISAGTIVAIVVPITVAVLIFIVGICFLSRRAR--KKQQGSVKEGKTAYDIPTV-DSL 788
Query: 340 ERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGK 399
+ F + ++ ATN FS D KLG+GGFG VYKG + + VAVK++S+ S QG
Sbjct: 789 Q-------FDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQV-VAVKRLSKSSGQGG 944
Query: 400 KEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG--KRTPLSWGV 457
+E+ EV V+++L+HRNLV+LLG+C E +LVYE++PN SLD LF K+ L WG
Sbjct: 945 EEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGR 1124
Query: 458 RHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
R+KI G+A G+ YLHE+ ++HRD+K+SN++LD
Sbjct: 1125RYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLD 1232
>TC218869 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (77%)
Length = 1309
Score = 149 bits (375), Expect = 4e-36
Identities = 76/153 (49%), Positives = 108/153 (69%)
Frame = +2
Query: 343 AGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEY 402
+G +++YKE+ AT NF+ LG+G FG VYK ++ VAVK + S+QG+KE+
Sbjct: 158 SGILKYSYKEIQKATQNFTNT--LGEGSFGTVYKAMMPTGEV-VAVKMLGPNSKQGEKEF 328
Query: 403 VTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIA 462
TEV ++ +L HRNLV LLG+C DKG+F+LVYEFM NGSL++ L+G+ LSW R +IA
Sbjct: 329 QTEVLLLGRLHHRNLVNLLGYCIDKGQFMLVYEFMSNGSLENLLYGEEKELSWDERLQIA 508
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
+ ++ G+ YLHE VVHRD+KS+N++LD S
Sbjct: 509 VDISHGIEYLHEGAVPPVVHRDLKSANILLDHS 607
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 149 bits (375), Expect = 4e-36
Identities = 87/226 (38%), Positives = 135/226 (59%), Gaps = 7/226 (3%)
Frame = +3
Query: 277 TLATSGDSKTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQK-----EEAMH 331
T+ +G+ K+ ++I++ V +I++ V Y + R RK+ N EE+
Sbjct: 3 TITYAGNKKSVSRVVLIVVPVVVSIILL---LCVCYFILKRSRKKYNTLLRENFGEESAT 173
Query: 332 LTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKI 391
L S+ +F ++ ATN FS ++++G+GGFG VYKG D ++AVKK+
Sbjct: 174 LESL-----------QFGLVTIEAATNKFSYEKRIGEGGFGVVYKGVLPD-GREIAVKKL 317
Query: 392 SRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG--K 449
S+ S QG E+ E+ +I++L+HRNLV LLG+C ++ E +L+YEF+ N SLD LF +
Sbjct: 318 SKSSGQGANEFKNEILLIAKLQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHR 497
Query: 450 RTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
L+W R+KI G+A G+ YLHE V+HRD+K SNV+LDS+
Sbjct: 498 SKQLNWSERYKIIEGIAQGISYLHEHSRLKVIHRDLKPSNVLLDSN 635
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 147 bits (371), Expect = 1e-35
Identities = 75/149 (50%), Positives = 105/149 (70%), Gaps = 2/149 (1%)
Frame = +3
Query: 347 RFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEV 406
+F + + ATNNFS KLGQGGFG VYKG +D ++A+K++S S QG+ E+ E+
Sbjct: 96 QFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSD-GQEIAIKRLSINSNQGETEFKNEI 272
Query: 407 KVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLF--GKRTPLSWGVRHKIALG 464
+ +L+HRNLV+LLG+C + E LL+YEF+PN SLD +F KR L+ +R+KI G
Sbjct: 273 LLTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFDPNKRVNLN*EIRYKIIRG 452
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A GLLYLHE+ VVHRD+K+SN++LD
Sbjct: 453 IARGLLYLHEDSRLNVVHRDLKTSNILLD 539
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 145 bits (365), Expect = 5e-35
Identities = 76/187 (40%), Positives = 121/187 (64%), Gaps = 13/187 (6%)
Frame = +2
Query: 322 SNKQKEEAMHLTSMND-DLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFA 380
+++ ++ +TS ++ ++ + + + F+Y EL AT NF D LG+GGFG+V+KG+
Sbjct: 224 NSRSSSASIPVTSRSEGEILQSSNLKSFSYHELRAATRNFRPDSVLGEGGFGSVFKGWID 403
Query: 381 DLDLQ---------VAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFL 431
+ L VAVKK+++ QG +E++ E+ + QL+H NLVKL+G+C + L
Sbjct: 404 EHSLAATKPGIGKIVAVKKLNQDGLQGHREWLAEINYLGQLQHPNLVKLIGYCFEDEHRL 583
Query: 432 LVYEFMPNGSLDSHLFGKRT---PLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSS 488
LVYEFMP GS+++HLF + + P SW +R KIALG A GL +LH E V++RD K+S
Sbjct: 584 LVYEFMPKGSMENHLFRRGSYFQPFSWSLRMKIALGAAKGLAFLHST-EHKVIYRDFKTS 760
Query: 489 NVMLDSS 495
N++LD++
Sbjct: 761 NILLDTN 781
>TC228411 similar to UP|Q94JZ6 (Q94JZ6) Protein kinase-like protein
(At3g24600), partial (28%)
Length = 751
Score = 144 bits (364), Expect = 7e-35
Identities = 73/153 (47%), Positives = 105/153 (67%), Gaps = 1/153 (0%)
Frame = +3
Query: 342 GAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKE 401
G FTY+EL AT+ FS LGQGGFG V++G + +VAVK++ GS QG++E
Sbjct: 15 GFSKSTFTYEELARATDGFSDANLLGQGGFGYVHRGILPN-GKEVAVKQLKAGSGQGERE 191
Query: 402 YVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTP-LSWGVRHK 460
+ EV++IS++ H++LV L+G+C + LLVYEF+PN +L+ HL GK P + W R +
Sbjct: 192 FQAEVEIISRVHHKHLVSLVGYCITGSQRLLVYEFVPNNTLEFHLHGKGRPTMDWPTRLR 371
Query: 461 IALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
IALG A GL YLHE+ ++HRDIK++N++LD
Sbjct: 372 IALGSAKGLAYLHEDCHPKIIHRDIKAANILLD 470
>BE801577 similar to PIR|T49986|T499 lectin-like protein kinase-like -
Arabidopsis thaliana, partial (7%)
Length = 421
Score = 143 bits (361), Expect = 2e-34
Identities = 72/139 (51%), Positives = 97/139 (68%), Gaps = 2/139 (1%)
Frame = -2
Query: 311 AYALFWRKRKRSNK--QKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQ 368
A+ + +KR+ +E+ H TS DL+R PRRF YKEL +ATN F+ D +LG+
Sbjct: 414 AWVIIMKKRRGKGDYYDNDESGH-TSAKFDLDRETIPRRFDYKELVVATNGFADDTRLGR 238
Query: 369 GGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKG 428
GG G VYKG + L VAVK+I S ++ ++ EV++IS+L HRNLV+ +GWCH++G
Sbjct: 237 GGSGQVYKGVLSHLGRVVAVKRIFTNSENSERVFINEVRIISRLIHRNLVQFVGWCHEQG 58
Query: 429 EFLLVYEFMPNGSLDSHLF 447
EFLLV+EFMPNGSLDSHLF
Sbjct: 57 EFLLVFEFMPNGSLDSHLF 1
>TC224358 similar to UP|Q8L7V7 (Q8L7V7) AT4g02010/T10M13_2, partial (26%)
Length = 688
Score = 143 bits (360), Expect = 2e-34
Identities = 74/155 (47%), Positives = 104/155 (66%), Gaps = 5/155 (3%)
Frame = +1
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTE 405
R Y+EL ATNNF LG+GGFG V+KG D VA+K+++ G +QG KE++ E
Sbjct: 139 RFIAYEELKEATNNFEPASVLGEGGFGRVFKGVLND-GTAVAIKRLTSGGQQGDKEFLVE 315
Query: 406 VKVISQLRHRNLVKLLGWC--HDKGEFLLVYEFMPNGSLDSHLFGK---RTPLSWGVRHK 460
V+++S+L HRNLVKL+G+ D + +L YE +PNGSL++ L G PL W R K
Sbjct: 316 VEMLSRLHHRNLVKLVGYYSNRDSSQNVLCYELVPNGSLEAWLHGPLGINCPLDWDTRMK 495
Query: 461 IALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
IAL A GL YLHE+ + CV+HRD K+SN++L+++
Sbjct: 496 IALDAARGLSYLHEDSQPCVIHRDFKASNILLENN 600
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 143 bits (360), Expect = 2e-34
Identities = 76/160 (47%), Positives = 105/160 (65%), Gaps = 10/160 (6%)
Frame = +1
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQ-------VAVKKISRGSRQG 398
R FT+ EL AT NF D LG+GGFG V+KG+ + +AVKK++ S QG
Sbjct: 106 RIFTFAELKAATRNFKADTVLGEGGFGKVFKGWLEEKATSKGGSGTVIAVKKLNSESLQG 285
Query: 399 KKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRT---PLSW 455
+E+ +EV + +L H NLVKLLG+C ++ E LLVYEFM GSL++HLFG+ + PL W
Sbjct: 286 LEEWQSEVNFLGRLSHTNLVKLLGYCLEESELLLVYEFMQKGSLENHLFGRGSAVQPLPW 465
Query: 456 GVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
+R KIA+G A GL +LH + V++RD K+SN++LD S
Sbjct: 466 DIRLKIAIGAARGLAFLHTSEK--VIYRDFKASNILLDGS 579
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 142 bits (358), Expect = 3e-34
Identities = 77/159 (48%), Positives = 103/159 (64%), Gaps = 10/159 (6%)
Frame = +1
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFAD---------LDLQVAVKKISRGSR 396
+ FT+ EL AT NF D LG+GGFG VYKG+ + + VAVKK+
Sbjct: 433 KAFTFNELKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGL 612
Query: 397 QGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK-RTPLSW 455
QG KE++TEV + QL H+NLVKL+G+C D LLVYEFM GSL++HLF + PLSW
Sbjct: 613 QGHKEWLTEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFRRGPQPLSW 792
Query: 456 GVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
VR K+A+G A GL +LH + V++RD K+SN++LD+
Sbjct: 793 SVRMKVAIGAARGLSFLHNAKSQ-VIYRDFKASNILLDA 906
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 142 bits (357), Expect = 4e-34
Identities = 73/147 (49%), Positives = 102/147 (68%), Gaps = 2/147 (1%)
Frame = +3
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVK 407
FT+ EL LAT FS+ L +GGFG+V++G D + +AVK+ S QG KE+ +EV+
Sbjct: 6 FTFSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQV-IAVKQYKLASTQGDKEFCSEVE 182
Query: 408 VISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLF-GKRTPLSWGVRHKIALGLA 466
V+S +HRN+V L+G+C D G LLVYE++ NGSLDSH++ K+ L W R KIA+G A
Sbjct: 183 VLSCAQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQNVLEWSARQKIAVGAA 362
Query: 467 SGLLYLHEEWE-RCVVHRDIKSSNVML 492
GL YLHEE C+VHRD++ +N++L
Sbjct: 363 RGLRYLHEECRVGCIVHRDMRPNNILL 443
>TC209710 similar to UP|Q9LPF9 (Q9LPF9) T3F20.25 protein, partial (19%)
Length = 566
Score = 142 bits (357), Expect = 4e-34
Identities = 75/159 (47%), Positives = 103/159 (64%), Gaps = 3/159 (1%)
Frame = +1
Query: 338 DLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQ 397
DL+ G FT +++ AT NF K+G+GGFG VYKG +D +AVK++S S+Q
Sbjct: 1 DLQTGL----FTLRQIKAATKNFDALNKIGEGGFGCVYKGQQSD-GTMIAVKQLSSKSKQ 165
Query: 398 GKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK---RTPLS 454
G +E+V E+ +IS L+H NLVKL G C + + +L+YE+M N L LFG+ +T L
Sbjct: 166 GNREFVNEMGLISGLQHPNLVKLYGCCVEGNQLILIYEYMENNCLSRILFGRDPNKTKLD 345
Query: 455 WGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
W R KI LG+A L YLHEE ++HRDIK+SNV+LD
Sbjct: 346 WPTRKKICLGIAKALAYLHEESRIKIIHRDIKASNVLLD 462
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 140 bits (352), Expect = 2e-33
Identities = 72/149 (48%), Positives = 104/149 (69%), Gaps = 2/149 (1%)
Frame = +2
Query: 347 RFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEV 406
RF + ++ AT FS+ KLG+GGFG VYKG +VAVK++S+ S QG +E+ EV
Sbjct: 41 RFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPS-GQEVAVKRLSKISGQGGEEFKNEV 217
Query: 407 KVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG--KRTPLSWGVRHKIALG 464
+++++L+HRNLV+LLG+C + E +LVYEF+ N SLD LF K+ L W R+KI G
Sbjct: 218 EIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVEG 397
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A G+ YLHE+ ++HRD+K+SNV+LD
Sbjct: 398 IARGIQYLHEDSRLKIIHRDLKASNVLLD 484
>TC229705 similar to UP|Q9LFP7 (Q9LFP7) Serine/threonine specific protein
kinase-like, partial (35%)
Length = 550
Score = 139 bits (351), Expect = 2e-33
Identities = 73/159 (45%), Positives = 104/159 (64%), Gaps = 10/159 (6%)
Frame = +1
Query: 336 NDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFAD---------LDLQV 386
+++L+ + R+FT+ EL LAT NF + LG+GGFG V+KG+ + L V
Sbjct: 70 SEELKVSSRLRKFTFNELKLATRNFRPESLLGEGGFGCVFKGWIEENGTAPVKPGTGLTV 249
Query: 387 AVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHL 446
AVK ++ QG KE++ E+ ++ L H NLVKL+G+C + + LLVYE MP GSL++HL
Sbjct: 250 AVKTLNHDGLQGHKEWLAELDILGDLVHPNLVKLVGFCIEDDQRLLVYECMPRGSLENHL 429
Query: 447 FGKRT-PLSWGVRHKIALGLASGLLYLHEEWERCVVHRD 484
F K + PL W +R KIALG A GL +LHEE +R V++RD
Sbjct: 430 FRKGSLPLPWSIRMKIALGAAKGLAFLHEEAQRPVIYRD 546
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 139 bits (351), Expect = 2e-33
Identities = 78/211 (36%), Positives = 121/211 (56%), Gaps = 38/211 (18%)
Frame = +1
Query: 321 RSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFA 380
R +K ++ ++ +DD A +F + + +AT +FS KLGQGGFGAVY+G +
Sbjct: 37 RRSKARKSSLVKQHEDDDEIEIAQSLQFNFDTIRVATEDFSDSNKLGQGGFGAVYRGRLS 216
Query: 381 DLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNG 440
D + +AVK++SR S QG E+ EV ++++L+HRNLV+LLG+C + E LL+YE++PN
Sbjct: 217 DGQM-IAVKRLSRESSQGDTEFKNEVLLVAKLQHRNLVRLLGFCLEGKERLLIYEYVPNK 393
Query: 441 SLDSHLFG--------------------------------------KRTPLSWGVRHKIA 462
SLD +FG K+ L+W +R+KI
Sbjct: 394 SLDYFIFGRS**VHN*TVALYFTSGSGQRLNIHIPLKLMAVYADPTKKAQLNWEMRYKII 573
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
G+A GLLYLHE+ ++HRD+K+SN++L+
Sbjct: 574 TGVARGLLYLHEDSHLRIIHRDLKASNILLN 666
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 139 bits (350), Expect = 3e-33
Identities = 83/211 (39%), Positives = 127/211 (59%), Gaps = 2/211 (0%)
Frame = +1
Query: 285 KTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAG 344
K+K++ +II T + +G ++A +R+ + ++++ + D+
Sbjct: 154 KSKKSSKIIIGTS----VAAPLGVVLAICFIYRRNIADKSKTKKSIDRQLQDVDVPL--- 312
Query: 345 PRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVT 404
F + AT+NF + K+G+GGFG VYKG ++AVK++S S QG E++T
Sbjct: 313 ---FDMLTITAATDNFLLNNKIGEGGFGPVYKGKLVG-GQEIAVKRLSSLSGQGITEFIT 480
Query: 405 EVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG--KRTPLSWGVRHKIA 462
EVK+I++L+HRNLVKLLG C E LLVYE++ NGSL+S +F K L W R I
Sbjct: 481 EVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRFNII 660
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
LG+A GLLYLH++ ++HRD+K+SNV+LD
Sbjct: 661 LGIARGLLYLHQDSRLRIIHRDLKASNVLLD 753
>TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase C, epsilon
type (nPKC-epsilon) , partial (5%)
Length = 908
Score = 139 bits (349), Expect = 4e-33
Identities = 77/172 (44%), Positives = 109/172 (62%), Gaps = 18/172 (10%)
Frame = +3
Query: 340 ERGAGPR--------RFTYKELDLATNNFSKDRKLGQGGFGAVYKGYF---------ADL 382
ERG P+ ++T EL AT NF D LG+GGFG V+KG+ +
Sbjct: 90 ERGERPQNNSVPKLIKYTLDELRSATRNFRPDTVLGEGGFGRVFKGWIDKNTFKPSRVGV 269
Query: 383 DLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSL 442
+ VAVKK + S QG +E+ +EV+++ + H NL KL+G+C ++ +FLLVYE+M GSL
Sbjct: 270 GIPVAVKKSNPDSLQGLEEWQSEVQLLGKFSHPNLGKLIGYCWEESQFLLVYEYMQKGSL 449
Query: 443 DSHLFGK-RTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+SHLF + PLSW +R KIA+G A GL +LH E+ V++RD KSSN++LD
Sbjct: 450 ESHLFRRGPKPLSWDIRLKIAIGAARGLAFLHTS-EKSVIYRDFKSSNILLD 602
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 138 bits (347), Expect = 6e-33
Identities = 78/163 (47%), Positives = 107/163 (64%), Gaps = 13/163 (7%)
Frame = +3
Query: 346 RRFTYKELDLATNNFSKDRKLG-QGGFGAVYKGYFADLDLQ---------VAVKKISRGS 395
+ F+ EL AT NF KD LG +G FG+V+KG+ + L VAVK++S S
Sbjct: 300 KNFSLTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKRLSLDS 479
Query: 396 RQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRT---P 452
QG K+ + EV + QL H +LVKL+G+C + + LLVYEFMP GSL++HLF + + P
Sbjct: 480 FQGHKDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLFMRGSYFQP 659
Query: 453 LSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
LSWG+R K+ALG A GL +LH E V++RD K+SNV+LDS+
Sbjct: 660 LSWGLRLKVALGAAKGLAFLHSA-ETKVIYRDFKTSNVLLDSN 785
>TC214696 similar to UP|Q8L683 (Q8L683) Lectin precursor, partial (80%)
Length = 1701
Score = 138 bits (347), Expect = 6e-33
Identities = 94/280 (33%), Positives = 147/280 (51%), Gaps = 10/280 (3%)
Frame = +3
Query: 10 KLIVSVETQSMASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAPRDG 69
K I ++ T + + L + F++ L+ ++ F + F+P NI+ Q A+
Sbjct: 24 KQIKAMATSNFSIVLSVSLAFFLVLLTKAHSTDTVSFTFNKFNPVQPNIMLQKDASISSS 203
Query: 70 EVN--FNINEN---YSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVINTQGRSPSLYG 124
V + N S +GR Y+ + +WDS TGK+ + T + F I +S S
Sbjct: 204 GVLQLTKVGSNGVPTSGSLGRALYAAPIQIWDSETGKVASWATSFKFNIFAPNKSNS--A 377
Query: 125 HGLAFFLVPYGFEIPLNSDGGFMGLFNTTTMVSSSNQIVHVEFDSFANREFREARGHVGI 184
GLAFFL P G + SD GF+GLFN+ + S Q V +EFD+F+N+++ A H+GI
Sbjct: 378 DGLAFFLAPVGSQP--QSDDGFLGLFNSP-LKDKSLQTVAIEFDTFSNKKWDPANRHIGI 548
Query: 185 NINSIISSATTPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTS--LSISI 242
++NSI S T W S +G AE+ + YN+ T L S +P + TS LS ++
Sbjct: 549 DVNSIKSVKTASWGLS--NGQVAEILVTYNAATSLLVAS-----LIHPSKKTSYILSDTV 707
Query: 243 DLMKVMPEWITVGFSAATSYVQ---ELNYLLSWEFNSTLA 279
+L +PEW++VGFSA T + E + ++SW F S L+
Sbjct: 708 NLKSNLPEWVSVGFSATTGLHEGSVETHDVISWSFASKLS 827
>BG352822 similar to GP|20453162|g At5g56885 {Arabidopsis thaliana}, partial
(13%)
Length = 471
Score = 137 bits (344), Expect = 1e-32
Identities = 70/140 (50%), Positives = 95/140 (67%), Gaps = 3/140 (2%)
Frame = +2
Query: 358 NNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNL 417
+NF R LG+GGFG VY G D +VAVK + R G +E+++EV+++S+L HRNL
Sbjct: 20 DNFHASRVLGEGGFGLVYSGTLED-GTKVAVKVLKREDHHGDREFLSEVEMLSRLHHRNL 196
Query: 418 VKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG---KRTPLSWGVRHKIALGLASGLLYLHE 474
VKL+G C + LVYE +PNGS++SHL G + +PL W R KIALG A GL YLHE
Sbjct: 197 VKLIGICAEVSFRCLVYELIPNGSVESHLHGVDKENSPLDWSARLKIALGSARGLAYLHE 376
Query: 475 EWERCVVHRDIKSSNVMLDS 494
+ V+HRD KSSN++L++
Sbjct: 377 DSSPHVIHRDFKSSNILLEN 436
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,783,589
Number of Sequences: 63676
Number of extensions: 305104
Number of successful extensions: 2792
Number of sequences better than 10.0: 654
Number of HSP's better than 10.0 without gapping: 2331
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2346
length of query: 495
length of database: 12,639,632
effective HSP length: 101
effective length of query: 394
effective length of database: 6,208,356
effective search space: 2446092264
effective search space used: 2446092264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146709.1


