

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146706.7 + phase: 0 /pseudo
(617 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
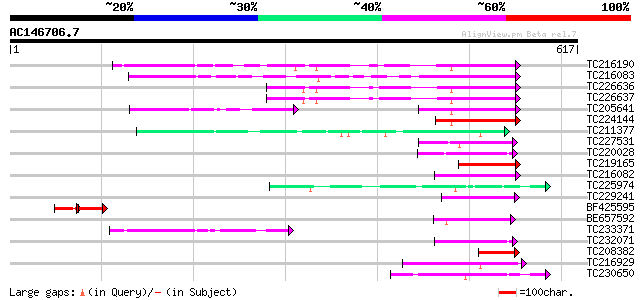
Score E
Sequences producing significant alignments: (bits) Value
TC216190 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ... 106 3e-23
TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 91 1e-18
TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL ... 91 2e-18
TC226637 UP|Q8S5C2 (Q8S5C2) 4-coumarate:CoA ligase isoenzyme 3 ... 89 7e-18
TC205641 UP|Q8S5C1 (Q8S5C1) 4-coumarate:CoA ligase isoenzyme 2 ... 85 1e-16
TC224144 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ... 82 5e-16
TC211377 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synth... 82 7e-16
TC227531 similar to UP|Q84P23 (Q84P23) 4-coumarate-CoA ligase-li... 80 2e-15
TC220028 weakly similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA li... 80 3e-15
TC219165 homologue to UP|4CL1_SOYBN (P31686) 4-coumarate--CoA li... 77 2e-14
TC216082 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 72 6e-13
TC225974 similar to UP|Q9SMT7 (Q9SMT7) 4-coumarate-CoA ligase-li... 71 2e-12
TC229241 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-li... 71 2e-12
BF425595 similar to GP|17065456|gb| A6 anther-specific protein {... 52 2e-11
BE657592 similar to GP|29888156|gb 4-coumarate-CoA ligase-like p... 66 4e-11
TC233371 weakly similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA li... 66 5e-11
TC232071 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 65 1e-10
TC208382 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-li... 60 2e-09
TC216929 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partia... 59 6e-09
TC230650 similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12... 58 1e-08
>TC216190 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
complete
Length = 1943
Score = 106 bits (265), Expect = 3e-23
Identities = 115/469 (24%), Positives = 193/469 (40%), Gaps = 25/469 (5%)
Frame = +3
Query: 112 KYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVAD 171
K+ D+ LID D T+TY ++ A A GL IG+ + I L N ++ +A
Sbjct: 138 KFHDRPCLIDG--DTGETLTYADVDLAARRIASGLHKIGIRQGDVIMLVLRNCPQFALAF 311
Query: 172 QGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVIL 231
G GA+ + EL + +++ L + +I D +S +++
Sbjct: 312 LGATHRGAVVTTANPFYTPAELAKQATATKT-RLVITQSAYVEKIKSFAD--SSSDVMVM 482
Query: 232 LWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYT 291
+ N+G ++H S A K I D++ L ++
Sbjct: 483 CIDDDFSYENDG-----------VLHFSTLSNADETEAPAVK------INPDELVALPFS 611
Query: 292 SGTTGNPKGVMLTHQNLL------------HQISKYAASLACI*KSL*VFHFF------- 332
SGT+G PKGVML+H+NL+ HQ + L C+ L +FH +
Sbjct: 612 SGTSGLPKGVMLSHKNLVTTIAQLVDGENPHQYTHSEDVLLCV---LPMFHIYALNSILL 782
Query: 333 --MRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGK 390
+R ++ +K E+ L + +++ +S V +V
Sbjct: 783 CGIRSGAAVLILQKFEITTLLELIEKYKVTVASFVPPIVL-------------------- 902
Query: 391 CLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSL 450
L K+ ++ Y D R + T P+ ++ V +++ A
Sbjct: 903 ALVKSGETHRY-----DLSSIRAVVTGAAPLGGELQEAVKARLPHA-------------- 1025
Query: 451 PSHVDRFFEAIGVTLQNGYGLTETSPVIA----ARRLSCNVIGSVGHPLKHTEFKVVDSE 506
T GYG+TE P+ A+ S G+ G +++ E K+VD+E
Sbjct: 1026-------------TFGQGYGMTEAGPLAISMAFAKEPSKIKPGACGTVVRNAEMKIVDTE 1166
Query: 507 TGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
TG+ LP G + +RG ++MKGY +P AT + IDK+GWL+TGDIG+I
Sbjct: 1167TGDSLPRNKSGEICIRGTKVMKGYLNDPEATERTIDKEGWLHTGDIGFI 1313
>TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (80%)
Length = 1411
Score = 91.3 bits (225), Expect = 1e-18
Identities = 115/435 (26%), Positives = 180/435 (40%), Gaps = 9/435 (2%)
Frame = +2
Query: 130 MTYKQLEDAILDFAEGLRV-IGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRS 188
+TY QL ++ A L V +G+ + + + NS + V +M+ GAI +
Sbjct: 161 LTYTQLWRSVEGVAASLSVDMGIRKGNVVLILSPNSIHFPVVCLAVMSLGAIITTTNPLN 340
Query: 189 SIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVP 248
+ E+ + S+ + LA ++ +I A + I+L N + +
Sbjct: 341 TTREIAKQIADSKPL-LAFTISDLLPKITAA-----APSLPIVLMDNDGANNNNNNNNI- 499
Query: 249 IFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNL 308
+ T E+ ++R V E ++ DD ATL+Y+SGTTG KGV
Sbjct: 500 VATLDEMAKKEPVAQR-----------VKERVEQDDTATLLYSSGTTGPSKGV------- 625
Query: 309 LHQISKYAASLACI*KSL*VFHFFMR----C*PSIYNCEKLEVYESLYSGIQRQISTSSL 364
+S + +A + L FH C +++ L + + G+ ST +
Sbjct: 626 ---VSSHRNLIAMVQIVLGRFHMEENETFICTVPMFHIYGLVAFAT---GLLASGSTIVV 787
Query: 365 VRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHML 424
+ K + L +E R T P L AML+ A +
Sbjct: 788 LSKFE----MHDMLSSIERFRA------TYLPLVPPILVAMLNNAAA------------I 901
Query: 425 AKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIAARR 482
K + +HS + SGG L V F A VT+ GYGLTE++ V A+
Sbjct: 902 KGKYDITSLHSVL-------SGGAPLSKEVIEGFVAKYPNVTILQGYGLTESTGVGASTD 1060
Query: 483 L--SCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQA 540
G+ G T+ +VD E+G+ LP G L +RGP +MKGY+ N AT
Sbjct: 1061SLEESRRYGTAGLLSPATQAMIVDPESGQSLPVNRTGELWLRGPTIMKGYFSNEEATTST 1240
Query: 541 IDKDGWLNTGDIGWI 555
+D GWL TGDIG+I
Sbjct: 1241LDSKGWLRTGDIGYI 1285
>TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL 2)
(4-coumaroyl-CoA synthase 2) (Clone 4CL16) , complete
Length = 1973
Score = 90.5 bits (223), Expect = 2e-18
Identities = 78/302 (25%), Positives = 129/302 (41%), Gaps = 26/302 (8%)
Frame = +1
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAAS------------LACI*KSL* 327
I DD + ++SGTTG PKGV+LTH++L +++ L C+ L
Sbjct: 619 IHPDDAVAMPFSSGTTGLPKGVILTHKSLTTSVAQQVDGENPNLYLTTEDVLLCV---LP 789
Query: 328 VFHFF---------MRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSL 378
+FH F +R ++ +K E+ L + ++S + +V LV
Sbjct: 790 LFHIFSLNSVLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVL-------- 945
Query: 379 GYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIG 438
L KN + A D R++ + P+ ++ + +++ A+
Sbjct: 946 ------------ALAKNP-----MVADFDLSSIRLVLSGAAPLGKELEEALRNRMPQAV- 1071
Query: 439 FSKAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGH 493
L GYG+TE PV++ A++ GS G
Sbjct: 1072--------------------------LGQGYGMTEAGPVLSMCLGFAKQPFQTKSGSCGT 1173
Query: 494 PLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG 553
+++ E KVVD ETG L G + +RG Q+MKGY + +AT ID +GWL+TGD+G
Sbjct: 1174VVRNAELKVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEAATASTIDSEGWLHTGDVG 1353
Query: 554 WI 555
++
Sbjct: 1354YV 1359
>TC226637 UP|Q8S5C2 (Q8S5C2) 4-coumarate:CoA ligase isoenzyme 3 , complete
Length = 1934
Score = 88.6 bits (218), Expect = 7e-18
Identities = 77/302 (25%), Positives = 128/302 (41%), Gaps = 26/302 (8%)
Frame = +1
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAAS------------LACI*KSL* 327
I+ DD + ++SGTTG PKGV+LTH++L +++ L C+ L
Sbjct: 610 IQPDDAVAMPFSSGTTGLPKGVVLTHKSLTTSVAQQVDGENPNLYLTTEDVLLCV---LP 780
Query: 328 VFHFF---------MRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSL 378
+FH F +R ++ +K E+ L + ++S + +V LV
Sbjct: 781 LFHIFSLNSVLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVL-------- 936
Query: 379 GYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIG 438
L KN + A D R++ + P+ + + +++ A+
Sbjct: 937 ------------ALAKNP-----MVADFDLSSIRLVLSGAAPLGKELVEALRNRVPQAV- 1062
Query: 439 FSKAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGH 493
L GYG+TE PV++ A++ GS G
Sbjct: 1063--------------------------LGQGYGMTEAGPVLSMCLGFAKQPFPTKSGSCGT 1164
Query: 494 PLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG 553
+++ E +VVD ETG L G + +RG Q+MKGY + AT ID +GWL+TGD+G
Sbjct: 1165VVRNAELRVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEKATALTIDSEGWLHTGDVG 1344
Query: 554 WI 555
++
Sbjct: 1345YV 1350
>TC205641 UP|Q8S5C1 (Q8S5C1) 4-coumarate:CoA ligase isoenzyme 2 , complete
Length = 1946
Score = 84.7 bits (208), Expect = 1e-16
Identities = 48/118 (40%), Positives = 65/118 (54%), Gaps = 7/118 (5%)
Frame = +1
Query: 445 SGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKH 497
SGG L ++ A L GYG+TE PV+ A+ G+ G +++
Sbjct: 997 SGGAPLGKELEDTLRAKFPNAKLGQGYGMTEAGPVLTMSLAFAKEPIDVKPGACGTVVRN 1176
Query: 498 TEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
E K+VD ETG LP G + +RG Q+MKGY + AT + IDKDGWL+TGDIG+I
Sbjct: 1177 AEMKIVDPETGHSLPRNQSGEICIRGDQIMKGYLNDGEATERTIDKDGWLHTGDIGYI 1350
Score = 48.5 bits (114), Expect = 9e-06
Identities = 45/184 (24%), Positives = 79/184 (42%)
Frame = +1
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
+Y +++ A GL+ GV + I + N ++ + G GA+ +
Sbjct: 226 SYHEVDSTARKVARGLKKEGVEQGQVIMILLPNCPEFVFSFLGASHRGAMATAANPFFTP 405
Query: 191 EELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIF 250
E+ + H+ + L + ++++ D+K + FV SC + K+ F
Sbjct: 406 AEIAK-QAHASNAKLLITQASYYDKVKDLRDIK--LVFV------DSCPPHTEEKQHLHF 558
Query: 251 TFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLH 310
+ L E + V IK DD+ L Y+SGTTG PKGVML+H+ L+
Sbjct: 559 S------------HLCEDNGDADVDVDVDIKPDDVVALPYSSGTTGLPKGVMLSHKGLVT 702
Query: 311 QISK 314
I++
Sbjct: 703 SIAQ 714
>TC224144 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (38%)
Length = 783
Score = 82.4 bits (202), Expect = 5e-16
Identities = 40/96 (41%), Positives = 61/96 (62%), Gaps = 4/96 (4%)
Frame = +2
Query: 464 TLQNGYGLTETSPVIA----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
T GYG+TE P+ A+ S G+ G +++ E K+VD+ETG+ LP G +
Sbjct: 314 TFGQGYGMTEAGPLAISMAFAKEPSKIKPGACGTVVRNAEMKIVDTETGDSLPRNKSGEI 493
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG ++MKGY +P AT + ID++GW++TGDIG+I
Sbjct: 494 CIRGAKVMKGYLNDPEATERTIDREGWVHTGDIGFI 601
>TC211377 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (58%)
Length = 1248
Score = 82.0 bits (201), Expect = 7e-16
Identities = 107/439 (24%), Positives = 172/439 (38%), Gaps = 33/439 (7%)
Frame = +1
Query: 139 ILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYN 198
+ FA G E++A FAD RW +A QG V S E L N
Sbjct: 1 VSSFASGXXSXXXRXXERVAXFADTRERWFIALQGCFRRNVTVVTMYSSLGKEALCHSLN 180
Query: 199 HSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIFTFTEIMHL 258
+E + E+ + + + L S++ VI + + + I TF+ + L
Sbjct: 181 ETEVTTVICGRKELKSLVNISGQLD-SVKRVICMDDDIPSDASSAQHGWKITTFSNVERL 357
Query: 259 GRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAAS 318
GR + S D+A ++YTSG+TG PKGVM+TH N+L +S S
Sbjct: 358 GR-----------ENPVEADLPLSADVAVIMYTSGSTGLPKGVMMTHGNVLATVS----S 492
Query: 319 LACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQI---------STSSLVRK-- 367
+ I +L ++ P + E V E+L + + I TS+ ++K
Sbjct: 493 VMIIVPNLGPKDVYLAYLPMAHILEL--VAENLIAAVGGCIGYGSPLTLTDTSNKIKKGK 666
Query: 368 -------------LVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLD----WLG 410
V RV G + K++ L+K +Y + W G
Sbjct: 667 QGDSTALMPTVMAAVPAILDRVRDGVL--KKVNSKGGLSKKLFHLAYSRRLQAINGCWFG 840
Query: 411 ARIIATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-AIGVTLQNG 468
A + L+ LV+ K+ + +G + + GG L RF +G + G
Sbjct: 841 AWGLEKALWNF------LVFKKVQAILGGRIRFILCGGAPLSGDTQRFINICLGAPIGQG 1002
Query: 469 YGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVL---PPGYKGILKVRGPQ 525
YGLTET + +G VG P+ + K++D G P +G + + GP
Sbjct: 1003YGLTETCAGGSFSDFDDTSVGRVGPPVPCSYIKLIDWPEGGYSTSDSPMARGEIVIGGPN 1182
Query: 526 LMKGYYKNPSATNQAIDKD 544
+ GY+KN T ++ D
Sbjct: 1183VTLGYFKNEEKTKESYKVD 1239
>TC227531 similar to UP|Q84P23 (Q84P23) 4-coumarate-CoA ligase-like protein,
partial (59%)
Length = 1478
Score = 80.5 bits (197), Expect = 2e-15
Identities = 51/113 (45%), Positives = 64/113 (56%), Gaps = 5/113 (4%)
Frame = +1
Query: 445 SGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIAARRLSCNVI---GSVGHPLKHTE 499
SGG L V F A V + GYGLTE+ AAR L + GSVG ++ E
Sbjct: 445 SGGAPLGKEVAEDFRAQFPNVEIGQGYGLTESGGG-AARVLGPDESKRHGSVGRLSENME 621
Query: 500 FKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDI 552
K+VD TGE L PG KG L +RGP +MKGY + AT + +D +GWL TGD+
Sbjct: 622 AKIVDPVTGEALSPGQKGELWLRGPTIMKGYVGDEKATAETLDSEGWLKTGDL 780
Score = 37.7 bits (86), Expect = 0.015
Identities = 15/36 (41%), Positives = 24/36 (66%)
Frame = +1
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKY 315
+ D A ++++SGTTG KGV+LTH+N + I +
Sbjct: 64 VSQSDSAAILFSSGTTGRVKGVLLTHRNFIALIGGF 171
>TC220028 weakly similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like
protein, partial (30%)
Length = 988
Score = 80.1 bits (196), Expect = 3e-15
Identities = 46/111 (41%), Positives = 63/111 (56%), Gaps = 2/111 (1%)
Frame = +3
Query: 444 ISGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFK 501
+ GG L D F+A V + GYGLTE S V N +G+ G + + E K
Sbjct: 129 VCGGSPLRKETDEAFKAKFPNVLVMQGYGLTE-SAVTRTTPEEANQVGATGKLIPNIEAK 305
Query: 502 VVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDI 552
+V+ ETGE + PG +G L +RGP +MKGY +P AT+ + DGWL TGD+
Sbjct: 306 IVNPETGEAMFPGEQGELWIRGPYVMKGYSGDPKATSATL-VDGWLRTGDL 455
>TC219165 homologue to UP|4CL1_SOYBN (P31686) 4-coumarate--CoA ligase 1 (4CL
1) (4-coumaroyl-CoA synthase 1) (Clone 4CL14) (Fragment)
, partial (48%)
Length = 425
Score = 77.0 bits (188), Expect = 2e-14
Identities = 34/67 (50%), Positives = 47/67 (69%)
Frame = +2
Query: 489 GSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLN 548
G+ G +++ E K+VD ETG LP + G + +RG Q+MKGY + AT + IDKDGWL+
Sbjct: 101 GACGTVVRNAELKIVDPETGHSLPRNHSGEICIRGDQIMKGYLNDGEATERTIDKDGWLH 280
Query: 549 TGDIGWI 555
TGDIG+I
Sbjct: 281 TGDIGYI 301
>TC216082 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (40%)
Length = 909
Score = 72.4 bits (176), Expect = 6e-13
Identities = 40/95 (42%), Positives = 54/95 (56%), Gaps = 2/95 (2%)
Frame = +1
Query: 463 VTLQNGYGLTETSPVIAARRL--SCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILK 520
VT+ GYGLTE++ V A+ G+ G T+ +VD E+G+ LP G L
Sbjct: 34 VTILQGYGLTESTGVGASTDSLEESRRYGTAGLLSPATQAMIVDPESGQSLPVNRTGELW 213
Query: 521 VRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RGP +MKGY+ N AT +D GWL TGDI +I
Sbjct: 214 LRGPTIMKGYFSNEEATTSTLDSKGWLRTGDICYI 318
>TC225974 similar to UP|Q9SMT7 (Q9SMT7) 4-coumarate-CoA ligase-like protein
(Adenosine monophosphate binding protein 3 AMPBP3),
partial (82%)
Length = 1836
Score = 70.9 bits (172), Expect = 2e-12
Identities = 79/314 (25%), Positives = 122/314 (38%), Gaps = 8/314 (2%)
Frame = +1
Query: 283 DDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KS-----L*VFHFFMRC*P 337
DD+A ++TSGTT PKGV LT NL ++ + L +FH
Sbjct: 529 DDVALFLHTSGTTSRPKGVPLTQHNLFSSVNNIKSVYRLTESDSTVIVLPLFHV------ 690
Query: 338 SIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQK 397
L +G+ + T + V A F S K
Sbjct: 691 -----------HGLIAGLLSSLGTGAAVALPAAGRFSASSFWKDMIK------------- 798
Query: 398 SPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRF 457
Y+ + I I+ H + + VY ++ S + P+ + +
Sbjct: 799 -----YSATWYTAVPTIHQIILDRHSNSPEPVYPRLRFIRSCSAS------LAPAILGKL 945
Query: 458 FEAIGVTLQNGYGLTETSPVIAARRL---SCNVIGSVGHPLKHTEFKVVDSETGEVLPPG 514
EA G + Y +TE S ++A+ L + GSVG P+ E ++D ETG V
Sbjct: 946 EEAFGAPVLEAYAMTEASHLMASNPLPQDGPHKAGSVGKPVGQ-EMVILD-ETGRVQDAE 1119
Query: 515 YKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIVVE 574
G + +RGP + KGY N A N A GW +TGD+G++ + G + +
Sbjct: 1120VSGEVCIRGPNVTKGYKNNVDA-NTAAFLFGWFHTGDVGYL----------DSDGYLHLV 1266
Query: 575 GRAKDTIVLSSEKV 588
GR K+ I EK+
Sbjct: 1267GRIKELINRGGEKI 1308
>TC229241 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-like protein,
partial (37%)
Length = 986
Score = 70.9 bits (172), Expect = 2e-12
Identities = 34/86 (39%), Positives = 52/86 (59%), Gaps = 2/86 (2%)
Frame = +3
Query: 471 LTETSPVIAAR--RLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMK 528
+TET +++ R+ GS G + E ++V +T + LPP G + VRGP +M+
Sbjct: 3 MTETCGIVSVENPRVGVRHTGSTGTLVSGVEAQIVSVDTQKPLPPRQLGEIWVRGPNMMQ 182
Query: 529 GYYKNPSATNQAIDKDGWLNTGDIGW 554
GY+ NP AT IDK GW++TGD+G+
Sbjct: 183 GYHNNPEATRLTIDKKGWVHTGDLGY 260
>BF425595 similar to GP|17065456|gb| A6 anther-specific protein {Arabidopsis
thaliana}, partial (3%)
Length = 221
Score = 52.4 bits (124), Expect(2) = 2e-11
Identities = 24/32 (75%), Positives = 25/32 (78%)
Frame = +3
Query: 75 LNPRFSPLLESSLLSGNDGVVSDEWKTVPDIW 106
L R SP LESSLLSGN GV S+EWK VPDIW
Sbjct: 126 LTRRCSPFLESSLLSGNGGVASNEWKAVPDIW 221
Score = 34.7 bits (78), Expect(2) = 2e-11
Identities = 19/30 (63%), Positives = 22/30 (73%)
Frame = +1
Query: 49 ILFLSSFLATASPLLSTFLELVSFANLNPR 78
+LFLSS LAT +P S F+ L SFAN NPR
Sbjct: 28 MLFLSSLLATTTPPGSIFIHL-SFANFNPR 114
>BE657592 similar to GP|29888156|gb 4-coumarate-CoA ligase-like protein
{Arabidopsis thaliana}, partial (35%)
Length = 780
Score = 66.2 bits (160), Expect = 4e-11
Identities = 39/92 (42%), Positives = 47/92 (50%), Gaps = 3/92 (3%)
Frame = -1
Query: 462 GVTLQNGYGLTE---TSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGI 518
GV +Q YGLTE + L SVG + E K VD +TG LP G
Sbjct: 720 GVAVQEAYGLTEHXXXXLTYVQKGLGSTNKNSVGFXXPNLEVKFVDPDTGRSLPRNTPGE 541
Query: 519 LKVRGPQLMKGYYKNPSATNQAIDKDGWLNTG 550
L VR +M+GYYK T Q IDK+GWL+TG
Sbjct: 540 LCVRSQCVMQGYYKQEDETAQTIDKNGWLHTG 445
>TC233371 weakly similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-like
protein, partial (45%)
Length = 943
Score = 65.9 bits (159), Expect = 5e-11
Identities = 57/203 (28%), Positives = 95/203 (46%), Gaps = 2/203 (0%)
Frame = +2
Query: 109 SAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWL 168
S + KIAL+D + T+T L+ + A G +G++ N+ + L A NS +
Sbjct: 158 SVSSFPSKIALVDSHSS--QTLTLAHLKSQVAKLAHGFLKLGINKNDVVLLLAPNSIHYP 331
Query: 169 VADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRF 228
+ A GA+ ++ E+ + + S L + PE+++++ K +L A
Sbjct: 332 ICFLAATAIGAVVSTXNPIYTVNEISKQVDDSNP-KLLITVPELWDKV-KNLNLPA---- 493
Query: 229 VILLWGEKSCLVN-EGSKEVPIFTFTE-IMHLGRGSRRLFESHDARKHYVFEAIKSDDIA 286
VI+ LV+ E EV T + +M + + L ES +K D A
Sbjct: 494 VIIDTETAQGLVSFEAGNEVSRITSLDAVMEMAGPATELPES----------GVKQGDTA 643
Query: 287 TLVYTSGTTGNPKGVMLTHQNLL 309
L+Y+SGTTG KGV+LTH+N +
Sbjct: 644 ALLYSSGTTGLSKGVVLTHRNFI 712
>TC232071 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (35%)
Length = 962
Score = 64.7 bits (156), Expect = 1e-10
Identities = 36/91 (39%), Positives = 51/91 (55%), Gaps = 1/91 (1%)
Frame = +2
Query: 463 VTLQNGYGLTE-TSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKV 521
V + GYGLTE T+ V N G+ G + E K+V+ TGE + P +G L +
Sbjct: 128 VMILQGYGLTESTAGVARTSPEDANRAGTTGRLVSGVEAKIVNPNTGEAMFPCEQGELWI 307
Query: 522 RGPQLMKGYYKNPSATNQAIDKDGWLNTGDI 552
+ P +MKGY +P AT+ + DGWL TGD+
Sbjct: 308 KSPSIMKGYVGDPEATSATL-VDGWLRTGDL 397
>TC208382 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-like protein,
partial (29%)
Length = 627
Score = 60.5 bits (145), Expect = 2e-09
Identities = 24/44 (54%), Positives = 33/44 (74%)
Frame = +3
Query: 511 LPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
LPPG G + VRGP +M+GY+ NP AT +DK GW++TGD+G+
Sbjct: 15 LPPGQLGEIWVRGPNMMQGYHNNPQATRLTMDKKGWVHTGDLGY 146
>TC216929 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial (51%)
Length = 1432
Score = 58.9 bits (141), Expect = 6e-09
Identities = 47/144 (32%), Positives = 65/144 (44%), Gaps = 9/144 (6%)
Frame = +1
Query: 428 LVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-AIGVTLQNGYGLTETSPVIAARRLSC 485
+V+ +I SA+G + + GG L F +G T+ GYGLTET
Sbjct: 265 IVFKQIRSALGGQLRFMLCGGAPLSGDSQHFINICMGATIGQGYGLTETFAGAVFSEWDD 444
Query: 486 NVIGSVGHPLKHTEFKVVDSETGEVL---PPGYKGILKVRGPQLMKGYYKNPSATNQA-- 540
+G VG PL K+V E G L P +G + V G + GY+KN T +
Sbjct: 445 YSVGRVGPPLPCCHIKLVSWEEGGYLTSDKPMPRGEIVVGGFSVTAGYFKNQEKTKEVFK 624
Query: 541 IDKDG--WLNTGDIGWIAAYHSSG 562
+D+ G W TGDIG +H G
Sbjct: 625 VDEKGMRWFYTGDIG---QFHPDG 687
>TC230650 similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12, partial
(43%)
Length = 1245
Score = 58.2 bits (139), Expect = 1e-08
Identities = 51/188 (27%), Positives = 88/188 (46%), Gaps = 14/188 (7%)
Frame = +3
Query: 415 ATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTE- 473
A I+F I + AK +I S++ ++GG P + E++G + + YG TE
Sbjct: 135 APIVFNIILEAKPSERIEIKSSVEI----LTGGAPPPPSLIDKIESLGFHVMHAYGSTEA 302
Query: 474 TSPVIAAR-RLSCNVIGSVGHP----------LKHTEFKVVDSETGEVLPPGYK--GILK 520
T P + + N + V L + V++ +T E +P K G +
Sbjct: 303 TGPALVCEWQQQWNQLPKVEQAQLKARQGISILTLEDVDVINVDTMESVPRDGKTMGEIV 482
Query: 521 VRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSGRSRNCGGVIVVEGRAKDT 580
+RG +MKGY K+P +T++A DGW +TGD+G + + G + ++ R+KD
Sbjct: 483 LRGSSIMKGYLKDPESTSKAF-CDGWFHTGDVGVV----------HKDGYLEIKDRSKDV 629
Query: 581 IVLSSEKV 588
I+ E +
Sbjct: 630 IISGGENI 653
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,793,971
Number of Sequences: 63676
Number of extensions: 416696
Number of successful extensions: 14536
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 5675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10987
length of query: 617
length of database: 12,639,632
effective HSP length: 103
effective length of query: 514
effective length of database: 6,081,004
effective search space: 3125636056
effective search space used: 3125636056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146706.7


