

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
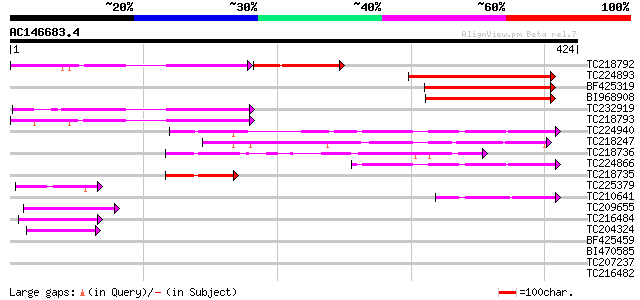
Query= AC146683.4 + phase: 0 /pseudo
(424 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218792 homologue to UP|O44991 (O44991) Lipid depleted protein ... 138 4e-60
TC224893 211 5e-55
BF425319 186 2e-47
BI968908 168 5e-42
TC232919 148 4e-36
TC218793 weakly similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial... 140 1e-33
TC224940 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein... 82 6e-16
TC218247 similar to UP|Q9FFV0 (Q9FFV0) Similarity to GTPase acti... 75 4e-14
TC218736 similar to UP|CV4L_HUMAN (Q9NU19) TBC1 domain family pr... 74 2e-13
TC224866 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein... 73 2e-13
TC218735 47 2e-05
TC225379 similar to UP|Q9AU06 (Q9AU06) NADPH-cytochrome P450 oxy... 45 6e-05
TC210641 weakly similar to UP|Q6PUR3 (Q6PUR3) C22orf4-like prote... 44 1e-04
TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 42 4e-04
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 42 5e-04
TC204324 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport fac... 41 9e-04
BF425459 similar to GP|21667485|gb| CONSTANS-like protein {Horde... 40 0.002
BI470585 40 0.002
TC207237 weakly similar to UP|EWS_HUMAN (Q01844) RNA-binding pro... 40 0.002
TC216482 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 40 0.003
>TC218792 homologue to UP|O44991 (O44991) Lipid depleted protein 6, partial
(4%)
Length = 756
Score = 138 bits (347), Expect(2) = 4e-60
Identities = 84/196 (42%), Positives = 105/196 (52%), Gaps = 15/196 (7%)
Frame = +3
Query: 1 MAKKRVPDWLNSPMWSSPTEQNR-FSATSYPSEPPSPPP--------VTVVE------DP 45
M KK+VPDWLNS +WSS + + SA + P E P PPP VT E P
Sbjct: 60 MVKKKVPDWLNSSLWSSSSPSSPPSSAVASPFEQPPPPPPRPPSPAVVTPSEPPPRPPSP 239
Query: 46 PSIHNAITTSPPSSSSSSASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQL 105
P++H+ + P S D + Q+QLLAE
Sbjct: 240 PAVHDPL---PTQRIDDSPREDHGDGFRSAHDVSRQAQLLAE------------------ 356
Query: 106 Y*YC*LFVY**LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELA 165
LS+KV+DM ELR +A QGIPD+ G+RST WKLLLGYLPPDR LWS+ELA
Sbjct: 357 -----------LSKKVVDMSELRSLACQGIPDAAGIRSTAWKLLLGYLPPDRGLWSAELA 503
Query: 166 KKRSQYKRFKQDILIN 181
KKRSQYK+FK++I +N
Sbjct: 504 KKRSQYKQFKEEIFMN 551
Score = 112 bits (279), Expect(2) = 4e-60
Identities = 52/68 (76%), Positives = 59/68 (86%)
Frame = +1
Query: 183 SEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQID 242
SEITR+MFNS + D D C R +LSRS+ITHGEHPLSLGKTS+WNQFFQDT+II+QID
Sbjct: 556 SEITRKMFNSTNCDTGDANC-ARALLSRSEITHGEHPLSLGKTSVWNQFFQDTEIIDQID 732
Query: 243 RDVKRTHP 250
RDVKRTHP
Sbjct: 733 RDVKRTHP 756
>TC224893
Length = 705
Score = 211 bits (537), Expect = 5e-55
Identities = 99/110 (90%), Positives = 106/110 (96%)
Frame = +1
Query: 299 VFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDE 358
VFKNDPDEENAAF+EAD FCFVELLSGFRDNF QQLDNS+VGIR+TITRLSQLL+EHDE
Sbjct: 1 VFKNDPDEENAAFAEADAVFCFVELLSGFRDNFVQQLDNSVVGIRATITRLSQLLREHDE 180
Query: 359 ELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
ELWRHLEVT+KVNPQFYAFRWITLLLTQEF+FADSL IWDTL+SDPDGPQ
Sbjct: 181 ELWRHLEVTSKVNPQFYAFRWITLLLTQEFNFADSLHIWDTLLSDPDGPQ 330
>BF425319
Length = 412
Score = 186 bits (471), Expect = 2e-47
Identities = 87/98 (88%), Positives = 93/98 (94%)
Frame = +1
Query: 311 FSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKV 370
F+EADTFFCFVELLS F+DNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKV
Sbjct: 1 FAEADTFFCFVELLSRFQDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKV 180
Query: 371 NPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
NPQFYAFRWI LLLTQEF+FAD L IWD ++SDP+GPQ
Sbjct: 181 NPQFYAFRWIILLLTQEFNFADILHIWDVILSDPEGPQ 294
>BI968908
Length = 394
Score = 168 bits (425), Expect = 5e-42
Identities = 78/97 (80%), Positives = 87/97 (89%)
Frame = -2
Query: 312 SEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVN 371
+EAD FFCFVELLSGFRDNF L NS+VGIR TITRLSQLL++HDEELWRHLEVT+KVN
Sbjct: 393 AEADAFFCFVELLSGFRDNFVHHLANSVVGIRXTITRLSQLLRDHDEELWRHLEVTSKVN 214
Query: 372 PQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
PQFYAFRWITLLLTQEF+FA +L WDTL++DP GPQ
Sbjct: 213 PQFYAFRWITLLLTQEFNFAATLXXWDTLLTDPHGPQ 103
>TC232919
Length = 457
Score = 148 bits (374), Expect = 4e-36
Identities = 91/182 (50%), Positives = 108/182 (59%), Gaps = 1/182 (0%)
Frame = +2
Query: 3 KKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSS 62
KK+VP+W+NS +WS+P S+PP+ V+V E+PP+I SPP SSSS
Sbjct: 5 KKQVPEWMNSSLWSTP------------SDPPAS--VSVREEPPTIPTNQNASPPLSSSS 142
Query: 63 SASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**LSRKVI 122
S S+ + ISR A QLLAE LSRKVI
Sbjct: 143 SVSSPTATADDISRHALFFVQLLAE-----------------------------LSRKVI 235
Query: 123 DMRELRKI-ASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILIN 181
DMRELR++ ASQGI D+ LR T+WKLLLGYLPPDRSLWSSEL KKRSQYK FK D+L N
Sbjct: 236 DMRELRRVVASQGISDAGALRPTLWKLLLGYLPPDRSLWSSELTKKRSQYKNFKDDLLTN 415
Query: 182 PS 183
PS
Sbjct: 416 PS 421
>TC218793 weakly similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (12%)
Length = 632
Score = 140 bits (353), Expect = 1e-33
Identities = 84/195 (43%), Positives = 101/195 (51%), Gaps = 12/195 (6%)
Frame = +3
Query: 1 MAKKRVPDWLNSPMWSS-----PTEQNRFSATSYPSEPPSPPPVTVVE-------DPPSI 48
M K +VPDWLNS +WSS P S P PPSP VT E PPS+
Sbjct: 144 MVKNKVPDWLNSSIWSSSSPSSPPSSAVASPFEQPPRPPSPAVVTPFEPPPPRPPSPPSV 323
Query: 49 HNAITTSPPSSSSSSASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*Y 108
H+ P S D + Q+QLLAE
Sbjct: 324 HDP---PPTQRIDDSPREDHGDGFRSAHDVSRQAQLLAE--------------------- 431
Query: 109 C*LFVY**LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKR 168
LS+KV+DM ELR +A QGIPD+ G+RST+WKLLLGYLPPDR LWS+ELAKKR
Sbjct: 432 --------LSKKVVDMSELRSLACQGIPDAAGIRSTVWKLLLGYLPPDRGLWSAELAKKR 587
Query: 169 SQYKRFKQDILINPS 183
QYK+FK++I +NPS
Sbjct: 588 FQYKQFKEEIFMNPS 632
>TC224940 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein-like,
partial (66%)
Length = 1783
Score = 81.6 bits (200), Expect = 6e-16
Identities = 70/295 (23%), Positives = 127/295 (42%), Gaps = 2/295 (0%)
Frame = +2
Query: 120 KVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAK--KRSQYKRFKQD 177
+V D LRK G D L++ +W LLLGY P + + E K K+ +Y+ K
Sbjct: 737 RVTDSEALRKRVFYGGLDHK-LQNEVWGLLLGYYPYESTYAEREFLKSVKKLEYENIKNQ 913
Query: 178 ILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDI 237
+S + + +F + +
Sbjct: 914 W---------------------------------------QSISSAQAKRFTKFRERKGL 976
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
IE +DV RT + F+ GD N L++IL+ ++ N + Y QGM+++L+P+
Sbjct: 977 IE---KDVVRTDRSLAFYEGDDN---PNVNVLRDILLTYSFYNFDLGYCQGMSDLLSPIL 1138
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
+V + E++ F+CFV L+ NF + + G+ S + LS+L++ D
Sbjct: 1139FVMDD----------ESEAFWCFVALMERLGPNFNRDQN----GMHSQLFALSKLVELLD 1276
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQVQIY 412
L + + +N F+ FRWI + +EF++ ++R+W+ L + + +Y
Sbjct: 1277SPLHNYFKQRDCLN-YFFCFRWILIQFKREFEYEKTMRLWEVLWTHYPSEHLHLY 1438
>TC218247 similar to UP|Q9FFV0 (Q9FFV0) Similarity to GTPase activating
protein, partial (77%)
Length = 1418
Score = 75.5 bits (184), Expect = 4e-14
Identities = 69/280 (24%), Positives = 120/280 (42%), Gaps = 19/280 (6%)
Frame = +2
Query: 145 IWKLLLGYLPPDRSLWSSELAK--KRSQYKRFKQDI-----LINPSE-ITRRMFNSASYD 196
+W+ LLG P + + + +R QY +K++ L+ +T +
Sbjct: 5 VWEFLLGCYDPKSTFEERDQIRQRRRMQYATWKEECHQLFPLVGSGRFVTAPVITEDGQP 184
Query: 197 ADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD--------IIEQIDRDVKRT 248
D ++ H ++ S N + TD + QI DV RT
Sbjct: 185 IQDPLVLKETSQAKGLAVHPQYNNSPSSMDAANNLAKVTDKTVVQWMLTLHQIGLDVVRT 364
Query: 249 HPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEEN 308
+ F+ L+K L +IL ++A ++ + Y QGM ++ +P+ + +
Sbjct: 365 DRTLVFYEKQENLSK-----LWDILAVYAWIDKDVGYGQGMCDLCSPMIILLDD------ 511
Query: 309 AAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTT 368
EAD F+CF L+ R NF + S VG+ + ++ L+ + + D +L +HLE
Sbjct: 512 ----EADAFWCFERLMRRLRGNF--RCTESSVGVAAQLSNLASVTQVIDPKLHKHLEHLG 673
Query: 369 KVNPQFYAFRWITLLLTQEFDFADSLRIWD---TLVSDPD 405
+AFR + +L +EF F DSL +W+ L DPD
Sbjct: 674 G-GDYLFAFRMLMVLFRREFSFCDSLYLWEMMWALEYDPD 790
>TC218736 similar to UP|CV4L_HUMAN (Q9NU19) TBC1 domain family protein
C6orf197, partial (23%)
Length = 1212
Score = 73.6 bits (179), Expect = 2e-13
Identities = 68/254 (26%), Positives = 114/254 (44%), Gaps = 13/254 (5%)
Frame = +2
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS V+ + +LR++A G+PD+ +R +W+LLLGY PP+ L +KR +Y
Sbjct: 464 LSGTVVILDKLRELAWSGVPDN--MRPKVWRLLLGYAPPNSDRREGVLRRKRLEYL---- 625
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
D + S YD D + RS + +
Sbjct: 626 DCI------------SQYYDIPDTE--------RSD--------------------DEVN 685
Query: 237 IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
++ QI D RT PD+ FF + Q++L+ IL +A +P YVQG+N+++ P
Sbjct: 686 MLHQIGIDCPRTVPDVPFF-----QQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 850
Query: 297 FYVFKN---DPDEENAAFS----------EADTFFCFVELLSGFRDNFCQQLDNSIVGIR 343
VF + + D N + S EAD ++C +LL G +D++ + GI+
Sbjct: 851 LVVFLSEHFEGDINNWSMSDLSSDIISNIEADCYWCLSKLLDGMQDHY----TFAQPGIQ 1018
Query: 344 STITRLSQLLKEHD 357
+ +L +L++ D
Sbjct: 1019RLVFKLKELVRRID 1060
>TC224866 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein-like,
partial (31%)
Length = 838
Score = 73.2 bits (178), Expect = 2e-13
Identities = 43/157 (27%), Positives = 81/157 (51%)
Frame = +3
Query: 256 CGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEAD 315
CGD N L++IL+ ++ N + Y QGM+++L+P+ +V N E++
Sbjct: 27 CGDDN---PNVNVLRDILLTYSFYNFDLGYCQGMSDLLSPILFVMDN----------ESE 167
Query: 316 TFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFY 375
F+CFV L+ NF + + G+ S + LS+L++ D L + + +N F+
Sbjct: 168 AFWCFVALMERLGPNFNRDQN----GMHSQLFALSKLVELLDSPLHNYFKQRDCLN-YFF 332
Query: 376 AFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQVQIY 412
FRWI + +EF++ ++R+W+ L + + +Y
Sbjct: 333 CFRWILIQFKREFEYEKTMRLWEVLWTHYPSEHLHLY 443
>TC218735
Length = 366
Score = 46.6 bits (109), Expect = 2e-05
Identities = 21/55 (38%), Positives = 35/55 (63%)
Frame = +3
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQY 171
LS ++ + +LR+ + +G+PD +R T+W+LLLGY PP+ L +KR +Y
Sbjct: 159 LSETMVKLEKLREFSWRGVPDY--MRPTVWRLLLGYAPPNSDRREGVLKRKRLEY 317
>TC225379 similar to UP|Q9AU06 (Q9AU06) NADPH-cytochrome P450 oxydoreductase
isoform 3, partial (13%)
Length = 737
Score = 45.1 bits (105), Expect = 6e-05
Identities = 29/69 (42%), Positives = 37/69 (53%), Gaps = 4/69 (5%)
Frame = +2
Query: 5 RVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTS----PPSSS 60
+ P SP+W S +R S+T P SPPP T PPS T+S PPS S
Sbjct: 404 KTPAQ*RSPLWISCPPSSRASSTP----PTSPPPPTPPPPPPSSSRTATSSCSSPPPSPS 571
Query: 61 SSSASTSST 69
SS+AS+SS+
Sbjct: 572 SSAASSSSS 598
>TC210641 weakly similar to UP|Q6PUR3 (Q6PUR3) C22orf4-like protein, partial
(14%)
Length = 707
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/95 (29%), Positives = 47/95 (49%), Gaps = 1/95 (1%)
Frame = +1
Query: 319 CFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFR 378
C +LL +D++ GI+ + +L +L++ D+ + H+E QF AFR
Sbjct: 1 CLSKLLDSMQDHYTFAQP----GIQRLVFKLKELVRRIDDPVSNHMEEQGLEFLQF-AFR 165
Query: 379 WITLLLTQEFDFADSLRIWDTLVSDPDG-PQVQIY 412
W LL +E F R+WDT +++ D P +Y
Sbjct: 166 WFNCLLIREIPFHLVTRLWDTYLAEGDALPDFLVY 270
>TC209655 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (32%)
Length = 832
Score = 42.4 bits (98), Expect = 4e-04
Identities = 25/72 (34%), Positives = 38/72 (52%)
Frame = +1
Query: 11 NSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTD 70
NSP ++P+ + + +S PS PP PP + N I+ SP SSS+ S+ST S
Sbjct: 169 NSPSPTAPSRLHSGTRSSSPSAPPPPPSPSRTPSFHGSRNTISASPTSSSTPSSSTPSAA 348
Query: 71 DMSISRLAQSQS 82
S ++ + S S
Sbjct: 349 PKSSTKPSYSPS 384
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 42.0 bits (97), Expect = 5e-04
Identities = 25/63 (39%), Positives = 31/63 (48%)
Frame = +1
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAST 66
P +SP SS T S+T P+ PPSPP T PP A +TSP S++ S S
Sbjct: 280 PSRTSSPSTSSSTTSAPRSSTRSPTAPPSPPQCTKPPAPPPDPPASSTSPTSTAEKSHSP 459
Query: 67 SST 69
T
Sbjct: 460 PKT 468
Score = 31.2 bits (69), Expect = 0.92
Identities = 25/76 (32%), Positives = 34/76 (43%)
Frame = +1
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAST 66
P SP S T + S T+ PS P SPP T PS A +T+PP + S ++
Sbjct: 103 PTRTTSPASSQSTPSSPPSTTTSPS-PTSPPKSTAKPPSPS---APSTTPPCRTFSRSTP 270
Query: 67 SSTDDMSISRLAQSQS 82
ST + S S +
Sbjct: 271 PSTPSRTSSPSTSSST 318
Score = 31.2 bits (69), Expect = 0.92
Identities = 16/50 (32%), Positives = 22/50 (44%)
Frame = +1
Query: 16 SSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAS 65
SSP R ++ + PS PP T P+ T PPS S+ S +
Sbjct: 88 SSPPSPTRTTSPASSQSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTT 237
Score = 29.3 bits (64), Expect = 3.5
Identities = 21/65 (32%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Frame = +1
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVE--DPPSIHNAITTSPPSSSSSSA 64
P SP +SP + + PS P + PP PPS + ++ SSS++SA
Sbjct: 154 PSTTTSPSPTSPPKST--AKPPSPSAPSTTPPCRTFSRSTPPSTPSRTSSPSTSSSTTSA 327
Query: 65 STSST 69
SST
Sbjct: 328 PRSST 342
>TC204324 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport factor 2
(NTF-2), complete
Length = 772
Score = 41.2 bits (95), Expect = 9e-04
Identities = 22/56 (39%), Positives = 31/56 (55%)
Frame = +2
Query: 13 PMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSS 68
P S ++R TS P+ PP P PPSI + ++PP SSSS+A++SS
Sbjct: 200 PCSLSRVRRSRAPTTSSPNSPPFPSNSASTPSPPSILSPQASTPPCSSSSAATSSS 367
>BF425459 similar to GP|21667485|gb| CONSTANS-like protein {Hordeum vulgare}
[Hordeum vulgare subsp. vulgare], partial (21%)
Length = 416
Score = 40.4 bits (93), Expect = 0.002
Identities = 20/55 (36%), Positives = 31/55 (56%)
Frame = +1
Query: 23 RFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDMSISRL 77
R +++ P+ PPS PP T PP+ A TT+ PSS S +T++T ++ L
Sbjct: 244 RRRSSARPTPPPSAPPATPTSTPPTPSPAATTASPSSPSPPPTTTTTTTTTLLTL 408
>BI470585
Length = 422
Score = 40.4 bits (93), Expect = 0.002
Identities = 18/40 (45%), Positives = 29/40 (72%)
Frame = +3
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPD 156
LS V+ + +LR++A G+PD+ +R +W+LLLGY PP+
Sbjct: 279 LSGTVVILDKLRELAWSGVPDN--MRPKVWRLLLGYAPPN 392
>TC207237 weakly similar to UP|EWS_HUMAN (Q01844) RNA-binding protein EWS
(EWS oncogene) (Ewing sarcoma breakpoint region 1
protein), partial (5%)
Length = 812
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/61 (40%), Positives = 34/61 (54%), Gaps = 2/61 (3%)
Frame = +3
Query: 16 SSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITT--SPPSSSSSSASTSSTDDMS 73
+S + +R SAT+ PS PP P T + P S +A ++ SP S S SSAST S S
Sbjct: 327 TSRSSASRSSATNSPSSPPPSWPTTTLPTPTSSSSATSSSRSPSSPSPSSASTPSPTSAS 506
Query: 74 I 74
+
Sbjct: 507 L 509
>TC216482 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (28%)
Length = 454
Score = 39.7 bits (91), Expect = 0.003
Identities = 24/58 (41%), Positives = 32/58 (54%), Gaps = 4/58 (6%)
Frame = +1
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVT----VVEDPPSIHNAITTSPPSSS 60
P +SP SS T S+T P+ PPSPPP T + DPP A++TSP S++
Sbjct: 292 PSRTSSPSTSSSTTSAPRSSTRSPTAPPSPPPCTRPPAALPDPP----ALSTSPTSTA 453
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,258,123
Number of Sequences: 63676
Number of extensions: 354509
Number of successful extensions: 10822
Number of sequences better than 10.0: 650
Number of HSP's better than 10.0 without gapping: 6076
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8857
length of query: 424
length of database: 12,639,632
effective HSP length: 100
effective length of query: 324
effective length of database: 6,272,032
effective search space: 2032138368
effective search space used: 2032138368
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146683.4


