

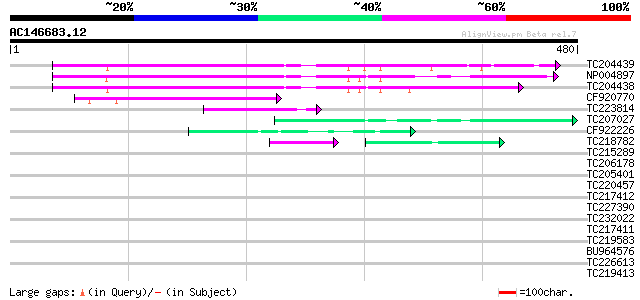
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.12 - phase: 0
(480 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 127 1e-29
NP004897 gag-protease polyprotein 125 4e-29
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 121 8e-28
CF920770 106 3e-23
TC223814 58 1e-08
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 50 2e-06
CF922226 45 7e-05
TC218782 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone n... 37 2e-04
TC215289 weakly similar to UP|Q9SJB0 (Q9SJB0) Expressed protein,... 38 0.009
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 35 0.057
TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {A... 35 0.057
TC220457 similar to UP|O24293 (O24293) Chloroplast inner envelop... 35 0.074
TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicas... 35 0.074
TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacety... 35 0.097
TC232022 homologue to UP|Q9LU46 (Q9LU46) DEAD-box protein abstra... 34 0.13
TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%) 34 0.13
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 33 0.22
BU964576 33 0.37
TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated pr... 32 0.48
TC219413 similar to UP|Q97LA8 (Q97LA8) Sensory transduction hist... 32 0.48
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 127 bits (319), Expect = 1e-29
Identities = 121/461 (26%), Positives = 200/461 (43%), Gaps = 31/461 (6%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 49 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++ I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 589 CNMRVDELIGSLQTFELGLSD-RAEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNK----LEYLARKQKKFLSK-----RGSYKNSKKEDQK-------GCF 315
DE + + +L + L + ++QK + R K K+ D K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCH 909
Query: 316 NCKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKFRKQIKK---SLMATWEDLDSESGSDK 372
C+ GH IA+CP K+ KG S S + S+ + +L +E + S +D
Sbjct: 910 GCEGYGHIIAECPTHLKKHRKGLSVCQSDTESEQESDSDRDVNALTGIFETAEDSSDTDS 1089
Query: 373 EEADDDAKAAMRLVATVSSEAVSEAE-------SDSEDENEVYSKIPRQELVDSLKELLS 425
E D+ A+ R + + SE + + E +D E E E + K EL + L S
Sbjct: 1090EITFDELAASYRKLC-IKSEKILQQEAQLKKVIADLEAEKEAH-KEEISELKGEVGFLNS 1263
Query: 426 LFEHRTNELTDLKEKYVDLMKQQKTTLLELKASEEELKGFN 466
E+ T + L + L + LL A + GFN
Sbjct: 1264KLENMTKSIKMLNKGSDTL---DEVLLLGKNAGNQRGLGFN 1377
>NP004897 gag-protease polyprotein
Length = 1923
Score = 125 bits (314), Expect = 4e-29
Identities = 117/450 (26%), Positives = 195/450 (43%), Gaps = 22/450 (4%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWD-ILEDGVDDLDLDEEGAAID----RRIHTPAQKKL 91
D + +WK M +F+ LD W +++D LD EG D T + +L
Sbjct: 49 DGTNYEYWKARMVAFLKSLDSRTWKAVIKDWEHPKMLDTEGKPTDGLKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++ I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
L V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 589 CNLRVDELIGSLQTFELGLSD-RTEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNK----LEYLARKQK------KFLSKRGSYKNSKKEDQK-------GC 314
DE + + +L + L + R+QK F ++GS + K+ D+K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKRSDEKPSHSKGFQC 906
Query: 315 FNCKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH A+CP K++ KG S S +D +SE SD +
Sbjct: 907 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESDSDR 1032
Query: 375 ADDDAKAAMRLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
A++ +ED ++ S+I EL S +EL E +
Sbjct: 1033---------------DVNALTGRFESAEDSSDTDSEITFDELATSYRELCIKSEKILQQE 1167
Query: 435 TDLKEKYVDLMKQQKTTLLELKASEEELKG 464
LK+ +L +++ E+ ELKG
Sbjct: 1168AQLKKVIANLEAEKEAHEEEI----SELKG 1245
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 121 bits (303), Expect = 8e-28
Identities = 107/426 (25%), Positives = 186/426 (43%), Gaps = 27/426 (6%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 49 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++ I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 589 CNMRVDELIGSLQTFELGLSD-RTEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNK----LEYLARKQK------KFLSKRGSYKNSKKEDQK-------GC 314
DE + + +L + L + R+QK F ++GS + KK D+K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKKSDEKPSHSKGIQC 906
Query: 315 FNCKKPGHFIADCPDLQKEKYKG----KSKKSSFSSSKFRKQIKKSLMATWEDLDSESGS 370
C+ GH A+CP K++ KG +S + + +L +E + S +
Sbjct: 907 HGCEGYGHIKAECPTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESAEDSSDT 1086
Query: 371 DKEEADDDAKAAMRLVATVSSEAV-SEAESDSEDENEVYSKIPRQELVDSLKELLSLFEH 429
D E D+ + R + S + + EA+ N K +E + LK +
Sbjct: 1087DSEITFDELAISYRELCIKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKGEVGFLNS 1266
Query: 430 RTNELT 435
+ +T
Sbjct: 1267KLENMT 1284
>CF920770
Length = 581
Score = 106 bits (264), Expect = 3e-23
Identities = 61/187 (32%), Positives = 102/187 (53%), Gaps = 12/187 (6%)
Frame = -2
Query: 56 DEELWDILEDG------VDDLDLDEEGAAIDRRIHTPAQK------KLYKKHHKIRGIIV 103
D +W+ +E G V+ + +D ++ I P + K + + K + II
Sbjct: 574 DLNIWEAIEIGPYIPTTVERVSIDGSSSSESITIEKPRDRWSEEDRKRVQYNLKAKNIIT 395
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDGSIEEM 163
+++ EY ++S+ +AK M+ +L EG+ VK ++ L H+YELFRM + +I+ M
Sbjct: 394 SALGMDEYFRVSNCKSAKEMWDTLRLTHEGTTDVKRSRINALTHEYELFRMNTNENIQSM 215
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
RF +V+ L L K + D ++K+LR L W+PKVTAI E++DL+ +S+ L L
Sbjct: 214 QKRFTHIVNHLAALGKEFQNEDLINKVLRCLSREWQPKVTAISESRDLSNMSLATLFGKL 35
Query: 224 KGHEMSL 230
+ HEM L
Sbjct: 34 QEHEMEL 14
>TC223814
Length = 607
Score = 57.8 bits (138), Expect = 1e-08
Identities = 40/101 (39%), Positives = 59/101 (57%), Gaps = 1/101 (0%)
Frame = +1
Query: 165 SRFQTLVSGLQILKKS-YVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
S+ Q +++ L+ L K+ DH++KIL+SL + RP V A+ ++KDL +L VE+ +L
Sbjct: 223 SKVQNIMNNLRSLSKT*DNHDDHITKILQSLLIQ*RP*VIALCDSKDLKSLPVEEFDGTL 402
Query: 224 KGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEE 264
+ HE+ L E E +K K IA SK KA K S S E
Sbjct: 403 QVHELELMEDEGQRKGKFIA-------SKVQKALKRSLSRE 504
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 50.1 bits (118), Expect = 2e-06
Identities = 58/258 (22%), Positives = 97/258 (37%), Gaps = 2/258 (0%)
Frame = +3
Query: 225 GHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSN 284
G + + + K K A KGK + ++E DGD +E + K
Sbjct: 216 GKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNKEKKDKEKGDGDGEEKKEKKDKEKEK 395
Query: 285 KLEYLAR-KQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSKKSS 343
K E + ++ L ++G KN + ED +G KK +KEK K KK
Sbjct: 396 KKEKKDKDEETDTLKEKG--KNDEGEDDEGNKKKKK--------DKKEKEKDHKKEKKDK 545
Query: 344 FSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAESDSED 403
K +++ S+ D+D E + E +D K E + + + +D
Sbjct: 546 EEGEKEDSKVEVSV----RDIDIEEIKKEGEKEDKGKDG-------GKEVKEKKKKEDKD 692
Query: 404 ENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELKASEEELK 463
+ E K+ ++ L L E ++ L EK D+ +Q K E EE K
Sbjct: 693 KKEKKKKVTGKDKTKDLSTLKQKLEKINGKIQPLLEKKADIERQIKEVEAEGHVVNEENK 872
Query: 464 GFNLIST-TYEDRLKILC 480
++ +D L + C
Sbjct: 873 N*GTVAVQDVQDTLILYC 926
>CF922226
Length = 667
Score = 45.1 bits (105), Expect = 7e-05
Identities = 49/193 (25%), Positives = 77/193 (39%), Gaps = 1/193 (0%)
Frame = -3
Query: 152 FRMKDDGSIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
F+M +D S+ E F L+ L+ + + D +L LP + + +D
Sbjct: 614 FKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYLPKSYSHFKETLLFGRD- 438
Query: 212 NTLSVEDLVSSLKGHEMSLNEHETSKKSKS-IALPSKGKTSKSSKAYKASESEEESPDGD 270
++S++++ ++L E LNE + K S S L ++GKT K D
Sbjct: 437 -SVSLDEVQTALNSKE--LNERKEKKSSASGEGLTARGKTFKK----------------D 315
Query: 271 SDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDL 330
S+ D+ +KQK K G K C++CKK GH CP+
Sbjct: 314 SEFDK---------------KKQKPENQKNGEGNIFKIR----CYHCKKEGHTRKVCPER 192
Query: 331 QKEKYKGKSKKSS 343
QK KK S
Sbjct: 191 QKNGGSNNRKKDS 153
>TC218782 similar to UP|Q41042 (Q41042) Pisum sativum L. (clone na-481-5),
partial (20%)
Length = 925
Score = 36.6 bits (83), Expect = 0.025
Identities = 33/109 (30%), Positives = 51/109 (46%), Gaps = 8/109 (7%)
Frame = +1
Query: 178 KKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKGHEMSLNEHETSK 237
KK AS S S +V ++ K++ ++ SS + S +E E K
Sbjct: 52 KKGKAASSSSSSDSSEDDSSDEDEVATKKQTKEVKVQKGKEESSS----DDSSSESEDEK 219
Query: 238 KSKSIALPSKGKTSK----SSKAYK----ASESEEESPDGDSDEDQSVK 278
+ +A+P K +++K S+ A K AS S ES D DSDED++ K
Sbjct: 220 PAAKVAVPPKNQSAKNGTLSTPAEKGKPAASSSSSESSDDDSDEDEAPK 366
Score = 32.7 bits (73), Expect(2) = 2e-04
Identities = 20/59 (33%), Positives = 30/59 (49%), Gaps = 1/59 (1%)
Frame = +1
Query: 221 SSLKGHEMSLNEHETSKKSKSIALPSKGKT-SKSSKAYKASESEEESPDGDSDEDQSVK 278
+S E S ++ + + KS P+ GK S+K + SES E D SDED++ K
Sbjct: 307 ASSSSSESSDDDSDEDEAPKSKVAPAAGKNVPASTKITQPSESSESDSDSSSDEDKNKK 483
Score = 30.0 bits (66), Expect(2) = 2e-04
Identities = 27/118 (22%), Positives = 43/118 (35%)
Frame = +3
Query: 302 SYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKFRKQIKKSLMATW 361
S+++ K D + + + D+ K K K S S K M
Sbjct: 525 SFESGKSSDDESSESSDEDNDAKPAVTDVSKPSALAKKKVDSSDSDDSSSDEDKGKM--- 695
Query: 362 EDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDS 419
D+D +S + EE ++ SSE SE ES+ E + E+VD+
Sbjct: 696 -DIDDDSSDESEEPQKKKTVKNPKESSDSSEEDSEDESEEEPSKTPQKRDRDVEMVDA 866
>TC215289 weakly similar to UP|Q9SJB0 (Q9SJB0) Expressed protein, partial
(32%)
Length = 1476
Score = 38.1 bits (87), Expect = 0.009
Identities = 30/103 (29%), Positives = 52/103 (50%), Gaps = 2/103 (1%)
Frame = +3
Query: 339 SKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLVATVSSEAVSEAE 398
S++SSF S + L+ ED D + ++EE DD+ KA R V + SS + S
Sbjct: 900 SRRSSFYS--WSNPQSMPLLPVDEDQDYDYEEEEEEEDDEEKA--RKVPSASSSSSSSLA 1067
Query: 399 SDSEDENEVYSKIPR--QELVDSLKELLSLFEHRTNELTDLKE 439
+ + E++V ++ R + ++ L F+ R+ L DL+E
Sbjct: 1068EEKKQEDQVQLRLNRVPESYAAHMRLRLGSFKARSFSLADLQE 1196
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 35.4 bits (80), Expect = 0.057
Identities = 15/45 (33%), Positives = 29/45 (64%), Gaps = 8/45 (17%)
Frame = +2
Query: 294 KKFLSKRGSYKNS--KKEDQKG------CFNCKKPGHFIADCPDL 330
+K S R SY+++ +++ ++G C NCK+PGH+ +CP++
Sbjct: 260 RKIRSDRFSYRDAPYRRDSRRGFSRDNLCKNCKRPGHYARECPNV 394
Score = 31.6 bits (70), Expect = 0.82
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +2
Query: 310 DQKGCFNCKKPGHFIADCPD 329
++K C NC+K GH DCP+
Sbjct: 638 NEKACNNCRKTGHLARDCPN 697
Score = 31.2 bits (69), Expect = 1.1
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = +2
Query: 314 CFNCKKPGHFIADCPD 329
C+NCK+PGH + CP+
Sbjct: 458 CWNCKEPGHMASSCPN 505
Score = 28.9 bits (63), Expect = 5.3
Identities = 22/89 (24%), Positives = 33/89 (36%), Gaps = 8/89 (8%)
Frame = +2
Query: 247 KGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNS 306
K K SS + S S SP D+ ++ S + R ++ S+ KN
Sbjct: 191 KKKWKMSSDSRSRSRSRSRSP-----MDRKIRSDRFSYRDAPYRRDSRRGFSRDNLCKNC 355
Query: 307 KKEDQKG--------CFNCKKPGHFIADC 327
K+ C NC PGH ++C
Sbjct: 356 KRPGHYARECPNVAICHNCGLPGHIASEC 442
>TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {Arabidopsis
thaliana;} , partial (16%)
Length = 912
Score = 35.4 bits (80), Expect = 0.057
Identities = 41/181 (22%), Positives = 71/181 (38%), Gaps = 20/181 (11%)
Frame = +3
Query: 217 EDLVSSLKGHEMSLNEHETSKKSK--SIALPSKGKTSKSSKAYKASESEEESPDGDSDED 274
ED+ K + S + E++KKSK IA+P+K ++ K S + +S D D D
Sbjct: 36 EDIKEKKKHSKTSSTKKESAKKSKIEKIAVPNKSRSPPKRAPKKPSSNLSKS---DEDSD 206
Query: 275 QSVKMAMLSNKLEYLARKQKKFLSKRGSY---------KNSKKEDQKG---------CFN 316
+S K+ K E +++ +K S K KKE + C
Sbjct: 207 ESPKVFSRKKKNEKGGKQKTATPTKSASEEKTEKVTRGKGKKKEKSRPSDNQLRDAICEI 386
Query: 317 CKKPGHFIADCPDLQKEKYKGKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEAD 376
K+ A D+ K+ K + + + I++ L ++ D E + E D
Sbjct: 387 LKEVNFNTATFTDILKKLAKQFDMDLTPRKASIKSMIQEELTKLADEADDEDREEDAEKD 566
Query: 377 D 377
+
Sbjct: 567 E 569
>TC220457 similar to UP|O24293 (O24293) Chloroplast inner envelope protein,
110 kD (IEP110) precursor, partial (32%)
Length = 1034
Score = 35.0 bits (79), Expect = 0.074
Identities = 40/145 (27%), Positives = 62/145 (42%), Gaps = 9/145 (6%)
Frame = +1
Query: 103 VASIPRTEYMKMSDKSTAKAMFASLCAN-FEGSKKVKEAKALMLVHQYELFRMKDDGSIE 161
VA++ R M + +A + +C + FE K VKEA A + DG
Sbjct: 304 VAALLRMRVMLCIPQQIVEAAHSDICGSLFE--KVVKEAIASGV-----------DGYDA 444
Query: 162 EMYSRFQTLVSGLQILKK--SYVASDHVSKILRSLPSRWRPKVTAIEEAKDL------NT 213
E+ + GL++ ++ +AS V KI + R R E AK+L NT
Sbjct: 445 EIQKSVRKAAHGLRLTREVAMSIASKAVRKIFINYIKRARAAGNRTESAKELKKMIAFNT 624
Query: 214 LSVEDLVSSLKGHEMSLNEHETSKK 238
L V +LV +KG ++ E K+
Sbjct: 625 LVVTNLVEDIKGESTDISSEEPVKE 699
>TC217412 similar to UP|Q6L724 (Q6L724) ATP-dependent RNA helicase, partial
(3%)
Length = 868
Score = 35.0 bits (79), Expect = 0.074
Identities = 25/96 (26%), Positives = 39/96 (40%), Gaps = 9/96 (9%)
Frame = +3
Query: 248 GKTSKSSKAYKASESEEESPDGDSDEDQSVKMA----MLSNKLEYLARKQK--KFL---S 298
G S+ + +K+S + GD DQS + N A K +L
Sbjct: 87 GSASRDRRGFKSSRGLDVEDSGDDFGDQSSRRGGRNFKTGNSWSRAAGKSSGDDWLIGGG 266
Query: 299 KRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEK 334
+R S +S CFNC + GH +DCP++ +
Sbjct: 267 RRSSRPSSSDRFGGTCFNCGESGHRASDCPNVSNRR 374
>TC227390 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (24%)
Length = 808
Score = 34.7 bits (78), Expect = 0.097
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Frame = +1
Query: 328 PDLQKEKYK-GKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMRLV 386
P+ + E+ K +SKK+ +S KQ+K ++ +D + +S D +E DD
Sbjct: 67 PETKAEELKVPESKKAVAKASASAKQVK--IVEPKKDNEEDSDDDSDEDDDFGS------ 222
Query: 387 ATVSSEAVSEAESDSEDENE 406
S E + +A+SDS+DE++
Sbjct: 223 ---SDEEMEDADSDSDDESD 273
>TC232022 homologue to UP|Q9LU46 (Q9LU46) DEAD-box protein abstrakt, partial
(39%)
Length = 829
Score = 34.3 bits (77), Expect = 0.13
Identities = 18/47 (38%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Frame = +2
Query: 312 KGCFNCKKPGHFIADCPDLQKEKYK--GKSKKSSFSSSKFRKQIKKS 356
KGC C GH I DCP L+ +K ++K F S +R +I K+
Sbjct: 569 KGCAYCGGLGHRIRDCPKLEHQKSMAIANNRKDYFGSGGYRGEI*KA 709
>TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%)
Length = 1275
Score = 34.3 bits (77), Expect = 0.13
Identities = 40/187 (21%), Positives = 71/187 (37%), Gaps = 15/187 (8%)
Frame = +3
Query: 158 GSIEEMYSRFQTLVSGLQILKKSYVAS---DHVSKILRSLPSRWRPKVTAIEEAKDLNTL 214
G + ++YS+ V + ++ V D +I + L +R P I + L L
Sbjct: 303 GFLSDVYSQAADEVGKIHLIADERVQGAVFDLPEEIAKELLNRDIPPGNTISKITKLPPL 482
Query: 215 SVEDLVSSLKGH--EMSLNEHETSKKSKSIALPSKGKTSKSSKAYKAS-----ESEEESP 267
+ S G + + S S+ G +S+ + +K+S E ++
Sbjct: 483 QDDGPPSDFYGRFSDRDRSSRRGSSTSRGGFSSRGGSSSRDRRGFKSSRGWDGEDSDDDD 662
Query: 268 DGDSDEDQSVKMAMLSNKLEYLARKQK--KFL---SKRGSYKNSKKEDQKGCFNCKKPGH 322
GD + + N A + +L S+R S +S CFNC + GH
Sbjct: 663 FGDRSSWRGGRNFKTGNSWSRAAGRSSGDDWLIGGSRRSSRPSSSDRFGGACFNCGESGH 842
Query: 323 FIADCPD 329
+DCP+
Sbjct: 843 RASDCPN 863
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 33.5 bits (75), Expect = 0.22
Identities = 11/20 (55%), Positives = 15/20 (75%)
Frame = +2
Query: 313 GCFNCKKPGHFIADCPDLQK 332
GCFNC + GHF +CP++ K
Sbjct: 542 GCFNCGEEGHFARECPNVGK 601
>BU964576
Length = 445
Score = 32.7 bits (73), Expect = 0.37
Identities = 27/112 (24%), Positives = 52/112 (46%), Gaps = 11/112 (9%)
Frame = +2
Query: 221 SSLKGHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDE---DQSV 277
++ + E++ E +K+K + SK + K ++ ESE + +GD+ E D S
Sbjct: 110 AAARAAEVAPPASEEKRKAKKAKVASKQQKKKEKESETVVESENKVEEGDASENVLDASN 289
Query: 278 KMAMLS-----NKLEYLARKQKKFLSKRG---SYKNSKKEDQKGCFNCKKPG 321
K ++ N + A + KK L K + + ++ +KGCF ++ G
Sbjct: 290 KTIVVEHCKQCNSFKTRANQVKKGLEKADIGITVILNPEKPRKGCFEIRQEG 445
>TC226613 similar to UP|Q8H0V8 (Q8H0V8) Spliceosome associated protein-like
(At4g21660), partial (59%)
Length = 1844
Score = 32.3 bits (72), Expect = 0.48
Identities = 31/136 (22%), Positives = 55/136 (39%), Gaps = 22/136 (16%)
Frame = +2
Query: 361 WEDLDSESGSDKEEADDDAK---------AAMRLVATVSSEAVSEAESDSEDENEVYSKI 411
W DL+ E ++EE +++ + A ++ V ++SS D D + K
Sbjct: 851 WGDLEEEEEEEEEEEEEEEEEEMEEEELEAGIQSVDSLSSTPTGVETPDVIDLRKQQRKE 1030
Query: 412 PRQELVDSLKE------------LLSLFEHRTNELTDLKEKYVDLMKQQKTTLLELKASE 459
P + L L+E + T K VDL++ QKT +++
Sbjct: 1031PERPLYQVLEEKEEKIAPGTLLGTTHTYVVGTGTQDKSGAKRVDLLRGQKTDKVDVTLQP 1210
Query: 460 EELKGF-NLISTTYED 474
EEL+ N++ YE+
Sbjct: 1211EELEAMENVLPAKYEE 1258
>TC219413 similar to UP|Q97LA8 (Q97LA8) Sensory transduction histidine
kinase, partial (4%)
Length = 953
Score = 32.3 bits (72), Expect = 0.48
Identities = 15/51 (29%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Frame = +1
Query: 231 NEHETSKKSKSIA-LPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMA 280
N+HE+ KK ++++ LP++GK K + +YK + ++ D +S + A
Sbjct: 154 NQHESEKKEENVSMLPNRGKDEKLTDSYKGRKKYKDVADAAQAAFESAEYA 306
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.126 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,324,708
Number of Sequences: 63676
Number of extensions: 185016
Number of successful extensions: 1159
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 1098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1150
length of query: 480
length of database: 12,639,632
effective HSP length: 101
effective length of query: 379
effective length of database: 6,208,356
effective search space: 2352966924
effective search space used: 2352966924
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146683.12


