

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.12 + phase: 0 /pseudo
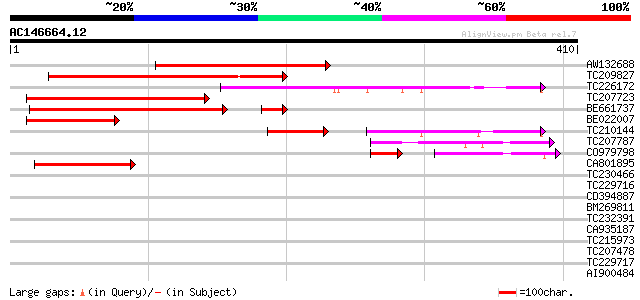
(410 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW132688 similar to GP|19715640|gb At1g47330/T3F24_2 {Arabidopsi... 169 2e-42
TC209827 158 4e-39
TC226172 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, part... 152 3e-37
TC207723 similar to UP|Q75LY7 (Q75LY7) Expressed protein, partia... 127 7e-30
BE661737 118 3e-29
BE022007 69 3e-12
TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, part... 66 3e-11
TC207787 58 9e-09
CO979798 40 1e-08
CA801895 55 6e-08
TC230466 similar to UP|Q8RY60 (Q8RY60) At1g47330/T3F24_2, partia... 33 0.23
TC229716 32 0.40
CD394887 similar to GP|11611669|gb| SLT1 protein {Arabidopsis th... 32 0.52
BM269811 weakly similar to GP|17381276|gb| AT4g33700/T16L1_190 {... 31 0.88
TC232391 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (14%) 29 4.4
CA935187 similar to GP|21555178|gb| transcription factor-like pr... 28 5.7
TC215973 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {A... 28 7.5
TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing pr... 28 7.5
TC229717 28 7.5
AI900484 homologue to GP|15163574|gb| AGR_pTi_174p {Agrobacteriu... 28 7.5
>AW132688 similar to GP|19715640|gb At1g47330/T3F24_2 {Arabidopsis thaliana},
partial (25%)
Length = 417
Score = 169 bits (429), Expect = 2e-42
Identities = 81/127 (63%), Positives = 104/127 (81%)
Frame = +1
Query: 106 AVLLATALISIIAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFPFAYPVSKLLDCLLGK 165
A+L++ LI + E++PQA+ +RYGL GAT++P VRVLL++FFP +YP+SK+LD +LGK
Sbjct: 34 AILISVTLILMFGEILPQAICTRYGLTVGATLAPLVRVLLIVFFPLSYPISKVLDWMLGK 213
Query: 166 GHTALLGREELKTLVNLHANEAGKGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSL 225
GH ALL R ELKT VN H NEAGKGG+LT ETTII GALDLT KTAKDAMTP+S+ FSL
Sbjct: 214 GHAALLKRAELKTFVNFHGNEAGKGGDLTHDETTIITGALDLTEKTAKDAMTPISKAFSL 393
Query: 226 DINSKLD 232
D+++ L+
Sbjct: 394 DLDATLN 414
>TC209827
Length = 761
Score = 158 bits (400), Expect = 4e-39
Identities = 81/173 (46%), Positives = 119/173 (67%)
Frame = +1
Query: 29 MFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVKNEHLVLCTLLMAKSL 88
+ FA I S L LGL+S VDLE+L ++G P + AA I+ +VK +H +L TLL+ ++
Sbjct: 244 VLFAGIMSGLTLGLMSLGLVDLEILERSGSPAEKMQAAIILPVVKKQHQLLVTLLLCNAV 423
Query: 89 ALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLRFGATMSPFVRVLLLLF 148
A+E + + ++K+F ++VA++L+ + EVIPQA+ SRYGL GA + VR+L+++
Sbjct: 424 AMEALPLYLDKLFNQFVAIILSVTFVLFFGEVIPQAICSRYGLAVGANFAWLVRILMIIC 603
Query: 149 FPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGELTLHETTII 201
+P +YPV K+LD LLG + AL R +LK LV++H EAGKGGELT ETTII
Sbjct: 604 YPVSYPVGKVLDHLLGH-NEALFRRAQLKALVSIHGQEAGKGGELTHDETTII 759
>TC226172 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, partial (62%)
Length = 1134
Score = 152 bits (383), Expect = 3e-37
Identities = 117/292 (40%), Positives = 156/292 (53%), Gaps = 57/292 (19%)
Frame = +3
Query: 153 YPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKGGELTLHETTIIAGALDLTMKTA 212
+P+SKLLD LLG H AL R ELKTLVNLH NEAGKGGELT ETTIIAGAL+L+ KTA
Sbjct: 3 FPISKLLDFLLGHRHEALFRRAELKTLVNLHGNEAGKGGELTHDETTIIAGALELSEKTA 182
Query: 213 KDAMTPLSETFSLDINSKLDM-----------SRI--------*CSAVRKMKHQLSL*L* 253
DAMTP+SETF++DINSKLD SR+ + +K+ L++
Sbjct: 183 SDAMTPISETFTVDINSKLDRELMNEILEKGHSRVPVYYEQPTNIIGLVLVKNLLTVHPE 362
Query: 254 GEFP------------GENWPLYDILNQFKKGQSHMAVVLK-------------SKENIR 288
E P E+ PLYDILN+F+KG SHMAVV++ + +++R
Sbjct: 363 DEAPVKSVTIRRIPRVPESMPLYDILNEFQKGHSHMAVVVRRCDKTNQQSSQNNANDSVR 542
Query: 289 TAATNTEG--------FGPFLP-HDYISI-STEASNWQSEGSEYYSATLKNAMLQESKDS 338
+ +G P +P H + S +T S+ + S +S + + +L+ D
Sbjct: 543 DVKVDIDGEKPPKEKALKPKMPLHKWKSFPNTNKSSNRGSRSRKWSKNMYSDILE--IDG 716
Query: 339 DPLHRSKQHDTSISLENMESLLGEEEVVGIITLEDVMEELLQVSL---TSYH 387
PL + L EEE VGIIT+EDV+EELLQ + T +H
Sbjct: 717 SPLPK---------------LPEEEEAVGIITMEDVIEELLQEEIFDETDHH 827
>TC207723 similar to UP|Q75LY7 (Q75LY7) Expressed protein, partial (33%)
Length = 492
Score = 127 bits (320), Expect = 7e-30
Identities = 60/132 (45%), Positives = 92/132 (69%)
Frame = +2
Query: 13 CCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
CC F+V ++ + + FA + S L LGL+S S VDLEVL K+G P +K+A KI+ +V
Sbjct: 95 CCSTAFFVHIVVIVLLVLFAGLMSGLTLGLMSLSLVDLEVLAKSGTPQDRKHAEKILPVV 274
Query: 73 KNEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLR 132
KN+HL+LCTLL+ + A+E + + ++ + W A+L++ LI + E+IPQ++ SRYGL
Sbjct: 275 KNQHLLLCTLLICNAAAMETLPIFLDGLVTAWGAILISVTLILLFGEIIPQSVCSRYGLA 454
Query: 133 FGATMSPFVRVL 144
GA+++PFVRVL
Sbjct: 455 IGASVAPFVRVL 490
>BE661737
Length = 793
Score = 118 bits (295), Expect(2) = 3e-29
Identities = 58/144 (40%), Positives = 94/144 (65%), Gaps = 1/144 (0%)
Frame = +2
Query: 15 GNHFW-VLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVK 73
G+ +W V A + + + FA I S L LGL+S VDLE+L ++G P +K AA I+ +V+
Sbjct: 119 GSVWWFVYAGISFFLVIFAGIMSGLTLGLMSLGLVDLEILQRSGSPSEKKQAAVILPVVQ 298
Query: 74 NEHLVLCTLLMAKSLALEGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRYGLRF 133
+H +L TLL+ + A+E + + ++KMF ++VA++L+ + EVIPQA+ SRYGL
Sbjct: 299 KQHQLLVTLLLCNAAAMEALPIYLDKMFNQYVAIILSVTFVLFFGEVIPQAICSRYGLAV 478
Query: 134 GATMSPFVRVLLLLFFPFAYPVSK 157
GA V +L+++++P AYP+ K
Sbjct: 479 GANFVWLVPILMIIWYPIAYPIWK 550
Score = 28.5 bits (62), Expect(2) = 3e-29
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = +3
Query: 183 HANEAGKGGELTLHETTII 201
H E GK GELT ETTII
Sbjct: 621 HXQEXGKXGELTHDETTII 677
>BE022007
Length = 332
Score = 69.3 bits (168), Expect = 3e-12
Identities = 32/67 (47%), Positives = 47/67 (69%)
Frame = +3
Query: 13 CCGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIV 72
CC F++ L+ + + FA + S L LGL+S S VDLEVL K+G P +K+AAKI+ +V
Sbjct: 132 CCETQFFLRLLIILLLVLFAGLMSGLTLGLMSLSLVDLEVLAKSGTPQDRKHAAKILPVV 311
Query: 73 KNEHLVL 79
+N+HL+L
Sbjct: 312 RNQHLLL 332
>TC210144 similar to UP|Q8VZI2 (Q8VZI2) AT4g33700/T16L1_190, partial (54%)
Length = 1167
Score = 65.9 bits (159), Expect = 3e-11
Identities = 32/44 (72%), Positives = 38/44 (85%)
Frame = +1
Query: 187 AGKGGELTLHETTIIAGALDLTMKTAKDAMTPLSETFSLDINSK 230
AGKGGELT ETTIIAGAL+L+ KTA DAMTP+++ FS+DIN K
Sbjct: 85 AGKGGELTHDETTIIAGALELSEKTASDAMTPITDIFSIDINFK 216
Score = 57.0 bits (136), Expect = 2e-08
Identities = 49/144 (34%), Positives = 69/144 (47%), Gaps = 15/144 (10%)
Frame = +2
Query: 259 ENWPLYDILNQFKKGQSHMAVVLKSKENIRTAATNTEG---------FGPFLPHD-YISI 308
E PLYDILN+F+KG SHMAVV++ E ++N G P + +
Sbjct: 395 ETLPLYDILNEFQKGHSHMAVVVRHCEKTGQQSSNNNADVRDVKVDIDGEKNPQENMLKT 574
Query: 309 STEASNWQSEGSEYYSATLKNAMLQESKD--SDPLHRSKQHDTSISLENMESLLGEEEVV 366
W+S + S + + SK+ SD L I ++ SL +EE V
Sbjct: 575 KRSLQKWKSFPNSNNSNRGGSRSRKWSKNMYSDIL--------EIDGNSLPSLPEKEEAV 730
Query: 367 GIITLEDVMEELLQVSL---TSYH 387
GIIT+EDV+EELLQ + T +H
Sbjct: 731 GIITMEDVIEELLQEEIFDETDHH 802
>TC207787
Length = 944
Score = 57.8 bits (138), Expect = 9e-09
Identities = 49/148 (33%), Positives = 66/148 (44%), Gaps = 15/148 (10%)
Frame = +3
Query: 262 PLYDILNQFKKGQSHMAVVLKSKENIRTAATNTEGFGPFLPHDYISISTEASNWQSEGSE 321
PLYDILN+F+KG SHMA V+K++ G G P E + S+
Sbjct: 168 PLYDILNEFQKGSSHMAAVVKAR-----------GKGKETPQIIDEEKNEENKSIGGDSQ 314
Query: 322 YYSATLK-------NAMLQESKDSDP--------LHRSKQHDTSISLENMESLLGEEEVV 366
+ L+ + ++ K S P L RS S EN+E + EV+
Sbjct: 315 LTTPLLQKQDAKSGSVVVDIVKPSKPSSINKLSVLQRSDSTTNGPSSENIE----DGEVI 482
Query: 367 GIITLEDVMEELLQVSLTSYHSTIIFVH 394
GIITLEDV EELLQ + + VH
Sbjct: 483 GIITLEDVFEELLQEEIVDETDEYVDVH 566
>CO979798
Length = 850
Score = 40.0 bits (92), Expect(2) = 1e-08
Identities = 34/95 (35%), Positives = 48/95 (49%), Gaps = 4/95 (4%)
Frame = -2
Query: 308 ISTEASNWQSEGSEYYSATLKNAMLQES-KDSDPLHRSKQHDTSISLENMESLLGEEEVV 366
++T Q+E SE A + S S L RS +N+E +EV+
Sbjct: 612 LTTPLLQKQNEMSENVVANIDKFSRPPSINKSTGLQRSDSRTNGSFSDNIE-----DEVI 448
Query: 367 GIITLEDVMEELLQVSLTS--YHSTI-IFVHLQTL 398
G+ITLEDV EELLQV+ ++ Y T+ VHL +L
Sbjct: 447 GVITLEDVFEELLQVNYSNLRYSPTLSTLVHLYSL 343
Score = 37.0 bits (84), Expect(2) = 1e-08
Identities = 14/23 (60%), Positives = 20/23 (86%)
Frame = -3
Query: 262 PLYDILNQFKKGQSHMAVVLKSK 284
P YDILN+F+KG SHMA V++++
Sbjct: 758 PXYDILNEFQKGSSHMAAVVRAR 690
>CA801895
Length = 454
Score = 55.1 bits (131), Expect = 6e-08
Identities = 29/73 (39%), Positives = 45/73 (60%)
Frame = +2
Query: 19 WVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVKNEHLV 78
+V A + + + FA I S L LGL+S VDLE+L + P +K AA I+ +V+ +H +
Sbjct: 101 FVYAGISFSLVIFAGIMSGLTLGLMSLGLVDLEILQSSCSPS*KKQAAVILPVVQKQHQL 280
Query: 79 LCTLLMAKSLALE 91
L TLL+ + A+E
Sbjct: 281 LVTLLLCNAAAME 319
Score = 35.8 bits (81), Expect = 0.036
Identities = 14/31 (45%), Positives = 22/31 (70%)
Frame = +2
Query: 202 AGALDLTMKTAKDAMTPLSETFSLDINSKLD 232
A A+++ + D+ +P+ TFSLD+NSKLD
Sbjct: 305 AAAMEIDCRRGHDSYSPIESTFSLDVNSKLD 397
>TC230466 similar to UP|Q8RY60 (Q8RY60) At1g47330/T3F24_2, partial (13%)
Length = 704
Score = 33.1 bits (74), Expect = 0.23
Identities = 15/32 (46%), Positives = 21/32 (64%)
Frame = +2
Query: 363 EEVVGIITLEDVMEELLQVSLTSYHSTIIFVH 394
E VVG+IT+EDV+EELLQ + + +H
Sbjct: 215 EVVVGVITMEDVIEELLQEEILDETDEYVNIH 310
>TC229716
Length = 839
Score = 32.3 bits (72), Expect = 0.40
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = -3
Query: 12 HCCGNHFWVLALLCWVFMF 30
HCC NH+W+L L + F F
Sbjct: 75 HCCSNHWWILVLAWFAFRF 19
>CD394887 similar to GP|11611669|gb| SLT1 protein {Arabidopsis thaliana},
partial (18%)
Length = 440
Score = 32.0 bits (71), Expect = 0.52
Identities = 12/36 (33%), Positives = 24/36 (66%)
Frame = +1
Query: 361 GEEEVVGIITLEDVMEELLQVSLTSYHSTIIFVHLQ 396
G+ E+ ++ LE+V+ L Q + ++HS +++ HLQ
Sbjct: 181 GKHELHALLLLEEVLAPLSQAQILAHHSMLLYWHLQ 288
>BM269811 weakly similar to GP|17381276|gb| AT4g33700/T16L1_190 {Arabidopsis
thaliana}, partial (14%)
Length = 420
Score = 31.2 bits (69), Expect = 0.88
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = +2
Query: 362 EEEVVGIITLEDVMEELLQ 380
+E VGIIT+EDV+EELLQ
Sbjct: 98 KEAAVGIITMEDVIEELLQ 154
>TC232391 similar to UP|Q7XA91 (Q7XA91) At3g27960, partial (14%)
Length = 562
Score = 28.9 bits (63), Expect = 4.4
Identities = 13/51 (25%), Positives = 24/51 (46%)
Frame = -1
Query: 103 EWVAVLLATALISIIAEVIPQALNSRYGLRFGATMSPFVRVLLLLFFPFAY 153
EWV+ ++ L+ ++ +P + N+ LRF ++ S F P Y
Sbjct: 322 EWVSRRVSNDLLDLLFRALPDSFNNSSNLRFSSSTSGFAVPSFSSLMPTTY 170
>CA935187 similar to GP|21555178|gb| transcription factor-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 450
Score = 28.5 bits (62), Expect = 5.7
Identities = 20/54 (37%), Positives = 23/54 (42%)
Frame = -1
Query: 137 MSPFVRVLLLLFFPFAYPVSKLLDCLLGKGHTALLGREELKTLVNLHANEAGKG 190
+SPFV L F PF V K L G REE K N + E G+G
Sbjct: 366 LSPFV-FFFLFFHPFQNEVPKYCSYLRTLGPRGHFSREE*KWYHNWRSGERGEG 208
>TC215973 similar to GB|AAP21204.1|30102572|BT006396 At3g15660 {Arabidopsis
thaliana;} , partial (70%)
Length = 797
Score = 28.1 bits (61), Expect = 7.5
Identities = 26/83 (31%), Positives = 37/83 (44%)
Frame = -1
Query: 14 CGNHFWVLALLCWVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVK 73
CGN V LL FF++ SS + L L S + + PH K+ M I+
Sbjct: 485 CGN---VGQLLKAFTAFFSSGSSKMFLALKGTSYCCNTLTARELNPHCGKSGTPFM*IIT 315
Query: 74 NEHLVLCTLLMAKSLALEGVSVL 96
L C++ AK + LE S+L
Sbjct: 314 GLSLTSCSITSAKEM-LEFSSLL 249
>TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing protein,
partial (51%)
Length = 1103
Score = 28.1 bits (61), Expect = 7.5
Identities = 13/36 (36%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Frame = -1
Query: 301 LPHDYIS---ISTEASNWQSEGSEYYSATLKNAMLQ 333
LPH ++S +S++ SNW + S+Y+S + +LQ
Sbjct: 200 LPHFHVSRDDLSSKESNWGTASSKYWSKAISLFILQ 93
>TC229717
Length = 427
Score = 28.1 bits (61), Expect = 7.5
Identities = 11/19 (57%), Positives = 13/19 (67%)
Frame = -3
Query: 13 CCGNHFWVLALLCWVFMFF 31
CC NH+WVL LL W + F
Sbjct: 74 CCSNHWWVL-LLAWFALRF 21
>AI900484 homologue to GP|15163574|gb| AGR_pTi_174p {Agrobacterium
tumefaciens str. C58 (Cereon)}, partial (2%)
Length = 370
Score = 28.1 bits (61), Expect = 7.5
Identities = 25/105 (23%), Positives = 47/105 (43%), Gaps = 1/105 (0%)
Frame = +3
Query: 26 WVFMFFAAISSALALGLLSFSQVDLEVLVKAGQPHIQKNAAKIMSIVKNEHLVLCTLLMA 85
WVF+ + +S Q+ +V +K G IQ+ + + +N LV+ +
Sbjct: 51 WVFLDLVEEQKRKSFVAVSAKQIWYDVRIKNGFEKIQEWTCNMKYLRRN--LVMEVINDY 224
Query: 86 KSLAL-EGVSVLMEKMFPEWVAVLLATALISIIAEVIPQALNSRY 129
SL + E +SVL ++FP ++ L + + P+A+ Y
Sbjct: 225 LSLTINEVISVLNLQLFPPTISSSLVEDEVEDVPMFCPEAVEQYY 359
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,444,693
Number of Sequences: 63676
Number of extensions: 221509
Number of successful extensions: 1882
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1865
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1878
length of query: 410
length of database: 12,639,632
effective HSP length: 100
effective length of query: 310
effective length of database: 6,272,032
effective search space: 1944329920
effective search space used: 1944329920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146664.12


