

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146664.11 + phase: 0
(248 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
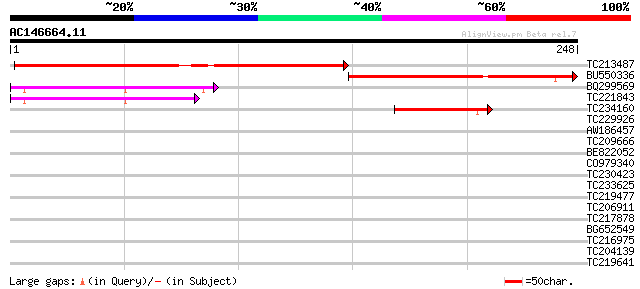
Score E
Sequences producing significant alignments: (bits) Value
TC213487 weakly similar to GB|AAP21375.1|30102914|BT006567 At3g6... 179 8e-46
BU550336 110 8e-25
BQ299569 70 9e-13
TC221843 67 8e-12
TC234160 similar to UP|AMRP_RAT (Q99068) Alpha-2-macroglobulin r... 44 9e-05
TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, part... 32 0.35
AW186457 weakly similar to GP|21109421|gb PilL protein {Xanthomo... 31 0.60
TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacu... 30 0.78
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 30 0.78
CO979340 30 1.0
TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fra... 30 1.3
TC233625 similar to UP|Q85DB9 (Q85DB9) NADH dehydrogenase subuni... 29 1.7
TC219477 similar to UP|Q8LSC5 (Q8LSC5) Geranylgeranyl pyrophosph... 28 3.0
TC206911 homologue to UP|Q9FK40 (Q9FK40) Histone acetyltransfera... 28 3.0
TC217878 similar to GB|AAQ65166.1|34365709|BT010543 At1g42550 {A... 28 3.9
BG652549 weakly similar to PIR|S62700|S62 photoassimilate-respon... 28 3.9
TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%) 28 5.1
TC204139 similar to UP|O04002 (O04002) CDSP32 protein (Chloropla... 28 5.1
TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 28 5.1
>TC213487 weakly similar to GB|AAP21375.1|30102914|BT006567 At3g61920
{Arabidopsis thaliana;} , partial (17%)
Length = 436
Score = 179 bits (455), Expect = 8e-46
Identities = 92/146 (63%), Positives = 109/146 (74%)
Frame = +2
Query: 3 NCLLGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPTNDQSSKPLCQFDE 62
NCLLG SDPDT+IK+ T +G IMEFY PITV+FITNEF GH IF ++D KPL QFDE
Sbjct: 2 NCLLGGMSDPDTIIKITTSNGCIMEFYAPITVSFITNEFPGHGIFLSHDLFCKPLSQFDE 181
Query: 63 LVAGQSYYLLPMTVLSPNNKIDYPSTCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRM 122
LVAGQSYYLLP+ NN+ ++PS A+ GG + IR GHVRS SVPTT P YRM
Sbjct: 182 LVAGQSYYLLPL-----NNQPEHPS--ASGHIGGDNRAIRLGHVRSHSVPTTHYPPTYRM 340
Query: 123 SLDYQYYQGVGLLKRTSTESFSCRTN 148
SLDYQ++QG+ L +TSTE FSCRT+
Sbjct: 341 SLDYQHHQGMRFLNKTSTEPFSCRTS 418
>BU550336
Length = 484
Score = 110 bits (274), Expect = 8e-25
Identities = 64/102 (62%), Positives = 75/102 (72%), Gaps = 2/102 (1%)
Frame = -2
Query: 149 RSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIESVRIVAKCGVT 208
RS + S+ S SS KS+ WKVKL ITP L++ILSQE TKELIESVRIVAKCGV
Sbjct: 477 RSRITXSSKRSKITSSSKSSRFWKVKLVITPXXLMDILSQEARTKELIESVRIVAKCGV- 301
Query: 209 AASSGGCGISSTTSIVSDQWSLSSSGRSA--PSKVDALVLDI 248
A+ G +S+ SIVSDQWSLSSSGRSA SK+DALV+D+
Sbjct: 300 -AAGGILPVSAAASIVSDQWSLSSSGRSACDSSKIDALVVDM 178
>BQ299569
Length = 423
Score = 70.1 bits (170), Expect = 9e-13
Identities = 41/97 (42%), Positives = 55/97 (56%), Gaps = 6/97 (6%)
Frame = +1
Query: 1 MGNCL---LGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPT-NDQSSKP 56
MGNC+ L D ++KV+T +G IME + PITV ITNEF G IF + D S+P
Sbjct: 34 MGNCVFRGLNHGVSEDMMVKVVTSNGGIMELFSPITVECITNEFPGQGIFRSRRDMFSEP 213
Query: 57 LCQFDELVAGQSYYLLPMTVLSPNNKI--DYPSTCAT 91
L + +EL G+ YYLLP+ S + + T AT
Sbjct: 214 LPKNEELHGGEVYYLLPLNPSSSRKSLTRQFSDTAAT 324
>TC221843
Length = 555
Score = 67.0 bits (162), Expect = 8e-12
Identities = 37/87 (42%), Positives = 52/87 (59%), Gaps = 4/87 (4%)
Frame = +3
Query: 1 MGNCL---LGANSDPDTLIKVITFDGNIMEFYPPITVNFITNEFQGHAIFPT-NDQSSKP 56
MGNC+ L + ++KV+T +G IME + PITV IT+EF GH IF + D S+P
Sbjct: 216 MGNCVFKGLHHGVSENMMVKVVTSNGGIMELFSPITVECITSEFPGHGIFRSRRDMFSEP 395
Query: 57 LCQFDELVAGQSYYLLPMTVLSPNNKI 83
L + +EL G+ YYLLP+ S +
Sbjct: 396 LPKNEELRGGKVYYLLPLNPSSSRKSL 476
>TC234160 similar to UP|AMRP_RAT (Q99068) Alpha-2-macroglobulin
receptor-associated protein precursor (Alpha-2-MRAP)
(Low density lipoprotein receptor-related
protein-associated protein 1) (RAP) (GP330-binding 45
kDa protein) (Fragment), partial (5%)
Length = 443
Score = 43.5 bits (101), Expect = 9e-05
Identities = 21/44 (47%), Positives = 31/44 (69%), Gaps = 1/44 (2%)
Frame = +1
Query: 169 GIWKVKLCITPEKLLEILSQEVSTKELIESVRIVA-KCGVTAAS 211
G+W+VKL I P +L +ILS++V+T+ LIE +R+ A C T S
Sbjct: 40 GVWRVKLVIDPRQLEKILSEQVNTEALIEKMRMAATACSTTTPS 171
>TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, partial (46%)
Length = 829
Score = 31.6 bits (70), Expect = 0.35
Identities = 28/100 (28%), Positives = 41/100 (41%)
Frame = +1
Query: 87 STCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCR 146
STC++A T S S P P P +L + G S S S R
Sbjct: 382 STCSSAATS------------SPSSAAAPPPTP--ATLPTRS*SGTTTCPAASASSPSAR 519
Query: 147 TNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEIL 186
+R+S + +T SSS +R S+ + C T + L+ L
Sbjct: 520 RSRASASAATESSSSSPTRSSSTTSPISKCCTKSRPLQTL 639
>AW186457 weakly similar to GP|21109421|gb PilL protein {Xanthomonas
axonopodis pv. citri str. 306}, partial (2%)
Length = 247
Score = 30.8 bits (68), Expect = 0.60
Identities = 24/80 (30%), Positives = 37/80 (46%)
Frame = -2
Query: 139 STESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEILSQEVSTKELIES 198
S S+S R RS ++ + S R + + +V L +P +L S T+
Sbjct: 237 SVTSWSWRPRRSRLSVRSPSEPRICASSDSSRERVMLT-SPARLTSRSSSGARTR----- 76
Query: 199 VRIVAKCGVTAASSGGCGIS 218
+ CG+T A SGGCGI+
Sbjct: 75 ----SGCGLTGARSGGCGIA 28
>TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacuolar thiol
protease precursor {Glycine max;} , partial (8%)
Length = 572
Score = 30.4 bits (67), Expect = 0.78
Identities = 35/109 (32%), Positives = 49/109 (44%), Gaps = 2/109 (1%)
Frame = +2
Query: 135 LKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLEI-LSQEVS-T 192
L + + SFS R++ SS + TIS S R S ST + +LL + LSQ +S
Sbjct: 173 LPSSPSHSFSSRSSSSSDFEFTISVSPRKS--STALCPADELFYKGQLLPLHLSQRISMV 346
Query: 193 KELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSAPSKV 241
+ L+ S + TA S G S++S SD S PS V
Sbjct: 347 RTLLLSSSSTSSTSTTARDSTG----SSSSFTSDLALFPDCDSSRPSSV 481
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis
thaliana}, partial (28%)
Length = 668
Score = 30.4 bits (67), Expect = 0.78
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +1
Query: 213 GGCGISSTTSIVSDQWSLSSSGRSAPS 239
GGCG SS++S S +S SSS S+PS
Sbjct: 256 GGCGTSSSSSSSSPSFSSSSSPSSSPS 336
>CO979340
Length = 747
Score = 30.0 bits (66), Expect = 1.0
Identities = 14/39 (35%), Positives = 22/39 (55%), Gaps = 5/39 (12%)
Frame = +2
Query: 143 FSCRTNRSSVNKS-----TISSSRRSSRKSTGIWKVKLC 176
F+C+ + V+K T+ + R RKS G WK+K+C
Sbjct: 470 FTCQIYKYCVSKQH*MHRTVPNKIRKIRKSKGKWKIKIC 586
>TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fragment),
partial (40%)
Length = 522
Score = 29.6 bits (65), Expect = 1.3
Identities = 12/31 (38%), Positives = 22/31 (70%)
Frame = -3
Query: 137 RTSTESFSCRTNRSSVNKSTISSSRRSSRKS 167
+ +T S +C T++++ + ST S+S+ SSR S
Sbjct: 208 KATTSSSTCTTSKATTSSSTCSTSKASSRSS 116
>TC233625 similar to UP|Q85DB9 (Q85DB9) NADH dehydrogenase subunit 4, partial
(5%)
Length = 890
Score = 29.3 bits (64), Expect = 1.7
Identities = 15/31 (48%), Positives = 21/31 (67%)
Frame = -2
Query: 138 TSTESFSCRTNRSSVNKSTISSSRRSSRKST 168
T + SFS T++ + ST+S+S RSS KST
Sbjct: 199 TVSASFSSSTDKCEESPSTVSASFRSSTKST 107
>TC219477 similar to UP|Q8LSC5 (Q8LSC5) Geranylgeranyl pyrophosphate
synthase, partial (77%)
Length = 1106
Score = 28.5 bits (62), Expect = 3.0
Identities = 21/66 (31%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Frame = +3
Query: 108 SQSVPTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCR-TNRSSVNKSTISSSRRSSRK 166
+Q P +P PAP R S + T+T + R RSS N S + RSSR
Sbjct: 399 AQRPPPSPPPAPSR*STPCRSSTTTCPAWTTTTSAVESRQATRSSANTWPSSPATRSSRS 578
Query: 167 STGIWK 172
+ W+
Sbjct: 579 RSSTWR 596
>TC206911 homologue to UP|Q9FK40 (Q9FK40) Histone acetyltransferase
(AT5g50320/MXI22_3), partial (61%)
Length = 1305
Score = 28.5 bits (62), Expect = 3.0
Identities = 20/65 (30%), Positives = 29/65 (43%)
Frame = +3
Query: 87 STCATAETGGGGQIIRQGHVRSQSVPTTPLPAPYRMSLDYQYYQGVGLLKRTSTESFSCR 146
+TC E G I+R+ HV +VP + D +QG G L E +CR
Sbjct: 765 TTCP--ELMGKCSIVRELHVYGTAVPV------HGRDADKLQHQGYGTLLMEEAERIACR 920
Query: 147 TNRSS 151
+RS+
Sbjct: 921 EHRST 935
>TC217878 similar to GB|AAQ65166.1|34365709|BT010543 At1g42550 {Arabidopsis
thaliana;} , partial (37%)
Length = 1685
Score = 28.1 bits (61), Expect = 3.9
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Frame = +2
Query: 155 STISSSRRSSRKSTGIWKV-KLCITPEKLLEILSQEVSTKELIESVRIVA 203
S +SS RR R +TG+W V + +T EKLL Q+V + +E+++I A
Sbjct: 647 SAMSSGRRE-RITTGLWNVEEEPLTAEKLLAFAMQKVESM-TVEALKIQA 790
>BG652549 weakly similar to PIR|S62700|S62 photoassimilate-responsive protein
PAR-1c precursor - common tobacco, partial (69%)
Length = 390
Score = 28.1 bits (61), Expect = 3.9
Identities = 14/50 (28%), Positives = 26/50 (52%)
Frame = +3
Query: 135 LKRTSTESFSCRTNRSSVNKSTISSSRRSSRKSTGIWKVKLCITPEKLLE 184
+KRT E+++CRT+ +K K+ G+ + L I+ + LL+
Sbjct: 171 VKRTGEEAYTCRTSEIEADKLKDHIETEQCIKACGLDRKSLGISSDSLLQ 320
>TC216975 similar to UP|Q6H974 (Q6H974) SEU1 protein, partial (28%)
Length = 1544
Score = 27.7 bits (60), Expect = 5.1
Identities = 14/54 (25%), Positives = 27/54 (49%)
Frame = +1
Query: 184 EILSQEVSTKELIESVRIVAKCGVTAASSGGCGISSTTSIVSDQWSLSSSGRSA 237
E+L + + ++ + V++ KC T A SG G+S + L++ G+ A
Sbjct: 88 ELLPRRLVAPQVNQLVQVAKKCQSTIAESGSDGVSQQDIQTNSNMLLTAGGQLA 249
>TC204139 similar to UP|O04002 (O04002) CDSP32 protein (Chloroplast
Drought-induced Stress Protein of 32kDa), partial (77%)
Length = 1243
Score = 27.7 bits (60), Expect = 5.1
Identities = 13/26 (50%), Positives = 17/26 (65%)
Frame = +3
Query: 146 RTNRSSVNKSTISSSRRSSRKSTGIW 171
RT SS + + SSSRRS+ TG+W
Sbjct: 255 RTRESSESTAWRSSSRRSNPPKTGLW 332
>TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 942
Score = 27.7 bits (60), Expect = 5.1
Identities = 14/27 (51%), Positives = 18/27 (65%)
Frame = -2
Query: 212 SGGCGISSTTSIVSDQWSLSSSGRSAP 238
+GGCG SS++S S S SSS S+P
Sbjct: 662 AGGCGTSSSSSSSSSSPSSSSSPSSSP 582
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.130 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,553,574
Number of Sequences: 63676
Number of extensions: 160779
Number of successful extensions: 768
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 754
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 764
length of query: 248
length of database: 12,639,632
effective HSP length: 95
effective length of query: 153
effective length of database: 6,590,412
effective search space: 1008333036
effective search space used: 1008333036
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146664.11


