

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
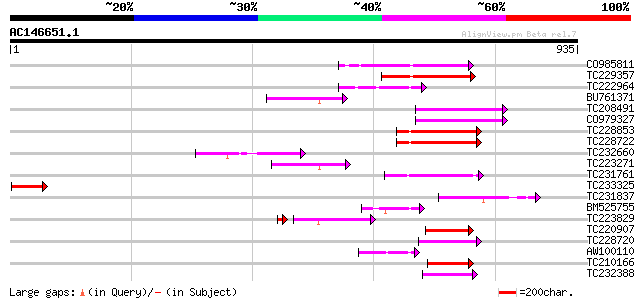
(935 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO985811 147 2e-35
TC229357 weakly similar to UP|Q8VDZ1 (Q8VDZ1) Hells protein, par... 144 1e-34
TC222964 similar to UP|Q925M9 (Q925M9) DNA-dependent ATPase SNF2... 107 2e-23
BU761371 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabi... 105 7e-23
TC208491 similar to UP|Q7XBI0 (Q7XBI0) Swi2/Snf2-related protein... 101 1e-21
CO979327 99 9e-21
TC228853 similar to UP|O04082 (O04082) Transcription factor RUSH... 98 2e-20
TC228722 weakly similar to UP|RAD5_YEAST (P32849) DNA repair pro... 97 3e-20
TC232660 weakly similar to UP|Q6M609 (Q6M609) Superfamily II DNA... 96 7e-20
TC223271 similar to UP|SN24_HUMAN (P51532) Possible global trans... 95 1e-19
TC231761 similar to UP|O23055 (O23055) YUP8H12.27 protein, parti... 87 3e-17
TC233325 86 6e-17
TC231837 similar to UP|ISWI_DROME (Q24368) Iswi protein (Imitati... 86 8e-17
BM525755 77 4e-14
TC223829 similar to UP|Q9LJK7 (Q9LJK7) DNA repair protein RAD54-... 70 1e-13
TC220907 weakly similar to UP|Q8NIR3 (Q8NIR3) Related to DNA rep... 74 3e-13
TC228720 weakly similar to UP|RAD5_YEAST (P32849) DNA repair pro... 72 1e-12
AW100110 weakly similar to GP|18463957|gb chromatin complex subu... 65 1e-10
TC210166 similar to UP|Q7XJP0 (Q7XJP0) SNF2/SWI2 family global t... 59 1e-08
TC232388 59 1e-08
>CO985811
Length = 713
Score = 147 bits (372), Expect = 2e-35
Identities = 94/226 (41%), Positives = 132/226 (57%), Gaps = 3/226 (1%)
Frame = -3
Query: 543 KTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYL--I 600
K ER++ E SS Y + ++ + + L +IG +S+ N VM+LR +CNHPYL +
Sbjct: 711 KIERLIRCEASS----YQKLLMKRVEENLGSIGNS-KXRSVHNSVMELRNICNHPYLSQL 547
Query: 601 PGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNI 660
E D+ + I+ KL +L +L L HRVL FS MT+LLD++E+YL
Sbjct: 546 HAEEVDNFIPKHYLPPIIRLCGKLEMLDRLLPKLKATDHRVLFFSTMTRLLDVMEEYLTS 367
Query: 661 EFGPKTYERVDGSVSVTDRQTAIARFNQDKSR-FVFLLSTRSCGLGINLATADTVIIYDS 719
+ Y R+DG S DR I FNQ S F+FLLS R+ G+G+NL ADTVI++D+
Sbjct: 366 K--QYRYLRLDGHTSGGDRGALIELFNQPGSPYFIFLLSIRAGGVGVNLQAADTVILFDT 193
Query: 720 DFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKL 765
D+NP D+QA RAHRIGQ +LV R +VEE++ A+ KL
Sbjct: 192 DWNPQVDLQAQARAHRIGQKRDVLVLRFETVQTVEEQVRASAEHKL 55
>TC229357 weakly similar to UP|Q8VDZ1 (Q8VDZ1) Hells protein, partial (34%)
Length = 1007
Score = 144 bits (364), Expect = 1e-34
Identities = 77/156 (49%), Positives = 104/156 (66%)
Frame = +2
Query: 613 LHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDG 672
L + + SAK L +L L + GHR LIFSQ T +LDILE L++ TY+R+DG
Sbjct: 71 LPDKHVMLSAKCRALAELLPSLKEGGHRALIFSQWTSMLDILEWTLDVI--GLTYKRLDG 244
Query: 673 SVSVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNR 732
S V +RQT + FN D S F LLSTR+ G G+NL ADTV+I+D DFNP D QA +R
Sbjct: 245 STQVAERQTIVDTFNNDTSIFACLLSTRAGGQGLNLTGADTVVIHDMDFNPQIDRQAEDR 424
Query: 733 AHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLD 768
HRIGQ+ + +YRLV + +V+E + ++AK+KL+LD
Sbjct: 425 CHRIGQTKPVTIYRLVTKGTVDENVYEIAKRKLVLD 532
>TC222964 similar to UP|Q925M9 (Q925M9) DNA-dependent ATPase SNF2H, partial
(10%)
Length = 422
Score = 107 bits (267), Expect = 2e-23
Identities = 64/145 (44%), Positives = 87/145 (59%)
Frame = +2
Query: 543 KTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPG 602
K E ++ V +S +Q +YYRA+L K+ ++ + G ++ +LNI MQLRK CNHPYL G
Sbjct: 8 KKETILKVGMSQMQKQYYRALLQKDLEV---VNAGGERKRLLNIAMQLRKCCNHPYLFQG 178
Query: 603 TEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEF 662
EP G + I+ + K+ LL +L L + RVLIFSQMT+LLDILEDYL F
Sbjct: 179 AEP--GPPFTTGDHLIENAGKMVLLDKLLPKLKERDSRVLIFSQMTRLLDILEDYL--MF 346
Query: 663 GPKTYERVDGSVSVTDRQTAIARFN 687
Y R+DG+ DR +I FN
Sbjct: 347 RGYQYCRIDGNTGGDDRDASIDAFN 421
>BU761371 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabidopsis
thaliana}, partial (19%)
Length = 451
Score = 105 bits (263), Expect = 7e-23
Identities = 59/149 (39%), Positives = 87/149 (57%), Gaps = 16/149 (10%)
Frame = +1
Query: 424 TSYEMVLADYS-HFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNN 482
TSYE+ L D +FR W+ ++VDEGHRLKNS+ KL L I+ ++++LLTGTPLQNN
Sbjct: 1 TSYEIALNDAKKYFRSYNWKYIVVDEGHRLKNSQCKLVKALKFINVENKLLLTGTPLQNN 180
Query: 483 LGEMYNLLNFLQPASFPSLSAFEERFN---------------DLTSAEKVDELKKLVSPH 527
L E+++LLNF+ P F SL FE FN + ++ V +L ++ P
Sbjct: 181 LAELWSLLNFILPDIFASLEEFESWFNLSGKCNNEATKEELEEKRRSQVVAKLHAILRPF 360
Query: 528 MLRRLKKDAMQNIPPKTERMVPVELSSIQ 556
+LRR+K D +P K E ++ ++ Q
Sbjct: 361 LLRRMKSDVEIMLPRKKEIIIYANMTEHQ 447
>TC208491 similar to UP|Q7XBI0 (Q7XBI0) Swi2/Snf2-related protein DDM1,
partial (22%)
Length = 839
Score = 101 bits (252), Expect = 1e-21
Identities = 62/153 (40%), Positives = 88/153 (56%), Gaps = 1/153 (0%)
Frame = +2
Query: 669 RVDGSVSVTDRQTAIARFNQDKSRF-VFLLSTRSCGLGINLATADTVIIYDSDFNPHADI 727
R+DG V + +R+ I FN S VFLLSTR+ GLGINL ADT I+YDSD+NP D+
Sbjct: 2 RIDGGVKLDERKQQIQDFNDVNSNCRVFLLSTRAGGLGINLTAADTCILYDSDWNPQMDL 181
Query: 728 QAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDILKW 787
QAM+R HRIGQ+ + VYRL S+E R+L+ A KL L+ + K +E
Sbjct: 182 QAMDRCHRIGQTKPVHVYRLSTAQSIEGRMLKRAFSKLKLEHVVIEKGQFHQERTKPASM 361
Query: 788 GTEELFNDSCALNGKDTSENNNSNKDEAVAEVE 820
E + L ++T+E+ + D + ++E
Sbjct: 362 DEIEEDDVLALLRDEETAEDKMIHTDISDEDLE 460
>CO979327
Length = 868
Score = 99.0 bits (245), Expect = 9e-21
Identities = 61/153 (39%), Positives = 86/153 (55%), Gaps = 1/153 (0%)
Frame = -1
Query: 669 RVDGSVSVTDRQTAIARFNQDKSRF-VFLLSTRSCGLGINLATADTVIIYDSDFNPHADI 727
R+DGSV +R+ I FN S V LSTR+ GLGINL ADT I+YDSD+NP D+
Sbjct: 859 RIDGSVKXXERKQQIQDFNDVNSNCRVXXLSTRAGGLGINLTVADTCILYDSDWNPQMDL 680
Query: 728 QAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKEVEDILKW 787
QAM+R HRIGQ+ + VYRL S+E R+L+ A KL L+ + K +E
Sbjct: 679 QAMDRCHRIGQTKPVHVYRLSTAQSIEGRMLKRAFSKLKLEHVVIEKGQFHQERTKPASM 500
Query: 788 GTEELFNDSCALNGKDTSENNNSNKDEAVAEVE 820
E + L ++T+E+ + D + ++E
Sbjct: 499 DEIEEDDVLALLRDEETAEDKKIHTDISDEDLE 401
>TC228853 similar to UP|O04082 (O04082) Transcription factor RUSH-1alpha
isolog; 18684-24052, partial (24%)
Length = 1393
Score = 97.8 bits (242), Expect = 2e-20
Identities = 56/140 (40%), Positives = 86/140 (61%)
Frame = +2
Query: 638 GHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLL 697
G + ++FSQ T++LDILE L + Y R+DG++SVT R A+ FN V ++
Sbjct: 710 GEKAIVFSQWTRMLDILEACL--KNSSIQYRRLDGTMSVTARDKAVKDFNTLPEVSVMIM 883
Query: 698 STRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERI 757
S ++ LG+N+ A V++ D +NP + QA++RAHRIGQ+ + V RL VR +VE+RI
Sbjct: 884 SLKAASLGLNMVAACHVLMLDLWWNPTTEDQAIDRAHRIGQTRPVTVLRLTVRDTVEDRI 1063
Query: 758 LQLAKKKLMLDQLFKGKSGS 777
L L +KK + G+ G+
Sbjct: 1064LALQQKKRTMVASAFGEDGT 1123
>TC228722 weakly similar to UP|RAD5_YEAST (P32849) DNA repair protein RAD5,
partial (4%)
Length = 1293
Score = 97.1 bits (240), Expect = 3e-20
Identities = 55/140 (39%), Positives = 85/140 (60%)
Frame = +3
Query: 638 GHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFVFLL 697
G + ++FSQ T++LD+LE L + Y R+DG++SV R A+ FN V ++
Sbjct: 510 GEKAIVFSQWTRMLDLLEACL--KNSSINYRRLDGTMSVVARDKAVKDFNTCPEVTVIIM 683
Query: 698 STRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERI 757
S ++ LG+NL A V++ D +NP + QA++RAHRIGQ+ + V RL VR +VE+RI
Sbjct: 684 SLKAASLGLNLVVACHVLMLDLWWNPTTEDQAIDRAHRIGQTRPVTVLRLTVRDTVEDRI 863
Query: 758 LQLAKKKLMLDQLFKGKSGS 777
L L +KK + G+ G+
Sbjct: 864 LDLQQKKRTMVASAFGEDGT 923
>TC232660 weakly similar to UP|Q6M609 (Q6M609) Superfamily II DNA/RNA
helicases, SNF2 family, partial (5%)
Length = 552
Score = 95.9 bits (237), Expect = 7e-20
Identities = 59/191 (30%), Positives = 96/191 (49%), Gaps = 10/191 (5%)
Frame = +1
Query: 307 LFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSR--------- 357
L HQ E + +L + + IL D+MGLGKTI A AF+++++ + S
Sbjct: 28 LLEHQREGVRFLYGLYKNNHGGILGDDMGLGKTIQAIAFLAAVFAKEGHSTLNENHVEKR 207
Query: 358 -PCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEA 416
P L++ P + NW +EF+ W+ + +V YHG + + + K EA
Sbjct: 208 DPALIICPTSVIHNWESEFSKWS-NFSVSIYHGANR---------------NLIYDKLEA 339
Query: 417 YKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTG 476
+ +L+TS++ S + W ++I+DE HRLKN +SKL+ I R LTG
Sbjct: 340 NEVEILITSFDTYRIHGSSLLDINWNIVIIDEAHRLKNEKSKLYKACLEIKTLRRYGLTG 519
Query: 477 TPLQNNLGEMY 487
T +QN + E++
Sbjct: 520 TAMQNKIMELF 552
>TC223271 similar to UP|SN24_HUMAN (P51532) Possible global transcription
activator SNF2L4 (SNF2-beta) (BRG-1 protein) (Mitotic
growth and transcription activator) (Brahma protein
homolog 1), partial (4%)
Length = 483
Score = 95.1 bits (235), Expect = 1e-19
Identities = 57/143 (39%), Positives = 85/143 (58%), Gaps = 13/143 (9%)
Frame = +3
Query: 432 DYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNS-ISFQHRVLLTGTPLQNNLGEMYNLL 490
D + + + W LIVDEGHRLKN E L L+S Q R+LLTGTP+QN+L E+++LL
Sbjct: 6 DKAFLKKIHWLYLIVDEGHRLKNHECALARTLDSGYHIQRRLLLTGTPIQNSLQELWSLL 185
Query: 491 NFLQPASFPSLSAFEERFN---------DLTSAEK---VDELKKLVSPHMLRRLKKDAMQ 538
NFL P F S+ FE+ FN LT E+ + L +++ P +LRR K + +
Sbjct: 186 NFLLPNIFNSVQNFEDWFNAPFADRVDVSLTDEEQLLIIRRLHQVIRPFILRRKKDEVEK 365
Query: 539 NIPPKTERMVPVELSSIQAEYYR 561
+P K++ ++ +LS+ Q YY+
Sbjct: 366 FLPSKSQVILKCDLSAWQKVYYQ 434
>TC231761 similar to UP|O23055 (O23055) YUP8H12.27 protein, partial (14%)
Length = 836
Score = 87.0 bits (214), Expect = 3e-17
Identities = 50/164 (30%), Positives = 91/164 (55%), Gaps = 2/164 (1%)
Frame = +2
Query: 619 KASAKLTLLHSMLKILYKE--GHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSV 676
+ S K+ L ++ + + + ++FSQ T LD++ ++ + +++GS+S+
Sbjct: 113 QTSTKIEALREEIRFMVERDGSAKGIVFSQFTSFLDLIN--YSLHKSGVSCVQLNGSMSL 286
Query: 677 TDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRI 736
R AI RF +D +FL+S ++ G+ +NL A V + D +NP + QA +R HRI
Sbjct: 287 AARDAAIKRFTEDPDCKIFLMSLKAGGVALNLTVASHVFLMDPWWNPAVERQAQDRIHRI 466
Query: 737 GQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKE 780
GQ + + R V+ ++EERIL+L +KK + +F+G G +
Sbjct: 467 GQYKPIRIVRFVIENTIEERILKLQEKK---ELVFEGTIGGSSD 589
>TC233325
Length = 347
Score = 86.3 bits (212), Expect = 6e-17
Identities = 46/61 (75%), Positives = 49/61 (79%), Gaps = 1/61 (1%)
Frame = +3
Query: 3 ENQTRLAEDNSACDNDLDGEIAENLVHDPQNVKSSD-EGELHNTDRVEKIHVYRRSITKE 61
ENQ+RL +DNSAC NDLD E EN + D QNVKSSD EG L NTDRVE IHVYRRSITKE
Sbjct: 165 ENQSRLLDDNSACANDLDVESTENHIDDCQNVKSSDEEGMLKNTDRVEGIHVYRRSITKE 344
Query: 62 S 62
S
Sbjct: 345 S 347
>TC231837 similar to UP|ISWI_DROME (Q24368) Iswi protein (Imitation swi
protein) (Nucleosome remodeling factor 140 kDa subunit)
(NURF-140) (CHRAC 140 kDa subunit), partial (13%)
Length = 788
Score = 85.9 bits (211), Expect = 8e-17
Identities = 60/177 (33%), Positives = 97/177 (53%), Gaps = 8/177 (4%)
Frame = +1
Query: 707 NLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLM 766
NLATA VI+YDSD+NP D+QA +RAHRIGQ + V+R ++EE++++ A KKL
Sbjct: 1 NLATAYVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTEYTIEEKVIERAYKKLA 180
Query: 767 LDQLF--KGKSGSQK-----EVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEV 819
LD L +G+ QK E+ ++++G E +F+ + + + + +EA AE+
Sbjct: 181 LDALVIQQGRLAEQKTVNKDELLQMVRFGAEMVFSSKDSTITDEDIDRIIAKGEEATAEL 360
Query: 820 EHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDAST-DIAEGDSEN 875
+ K + K T+++ K D+ A +L D + +D S DI + SEN
Sbjct: 361 DAKMK------------KFTEDAIKFKMDDTA--ELYDFDDEKDESRFDIKKIVSEN 489
>BM525755
Length = 457
Score = 77.0 bits (188), Expect = 4e-14
Identities = 46/107 (42%), Positives = 65/107 (59%), Gaps = 4/107 (3%)
Frame = +3
Query: 581 QSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMR----IKASAKLTLLHSMLKILYK 636
+++ N M+LRK CNHP L + L E+ +K+ KL +L +L L +
Sbjct: 162 KTLNNRCMELRKTCNHPSL---------NFPLLSELSTNSIVKSCGKLWILDRILIKLQR 314
Query: 637 EGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAI 683
GHRVL+FS MTKLLD+LEDYLN + Y R+DG+ S+ DR++AI
Sbjct: 315 TGHRVLLFSTMTKLLDLLEDYLN--WRRLVYRRIDGTTSLDDRESAI 449
>TC223829 similar to UP|Q9LJK7 (Q9LJK7) DNA repair protein RAD54-like,
partial (22%)
Length = 679
Score = 70.5 bits (171), Expect(2) = 1e-13
Identities = 44/150 (29%), Positives = 75/150 (49%), Gaps = 15/150 (10%)
Frame = +3
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERF---------------NDLTS 513
+ R+LL+GTPLQN+L E + ++NF P ++ F +
Sbjct: 141 KRRILLSGTPLQNDLEEFFAMVNFTNPGILGDIAHFRRYYEAPIICGREPAATAEEKKLG 320
Query: 514 AEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRN 573
AE+ EL V+ +LRR ++PPK +V +L+ +Q+E Y+ + ++ + R
Sbjct: 321 AEQSAELSVNVNRFILRRTNALLSNHLPPKIVEVVCCKLTPLQSELYKHFI-QSKNVKRA 497
Query: 574 IGKGIAQQSMLNIVMQLRKVCNHPYLIPGT 603
I + + Q +L + L+K+CNHP LI T
Sbjct: 498 ITEELKQSKILAYITALKKLCNHPKLIYDT 587
Score = 24.6 bits (52), Expect(2) = 1e-13
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = +2
Query: 442 EVLIVDEGHRLKNSES 457
++LI DE HRLKN ++
Sbjct: 35 DLLICDEAHRLKNDQT 82
>TC220907 weakly similar to UP|Q8NIR3 (Q8NIR3) Related to DNA repair protein
RAD26, partial (10%)
Length = 962
Score = 73.9 bits (180), Expect = 3e-13
Identities = 36/82 (43%), Positives = 58/82 (69%), Gaps = 2/82 (2%)
Frame = +1
Query: 686 FNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVY 745
FN S+ VFL+STR+ GLG+NL +A+ V+I+D ++NP D+QA +R+ R GQ ++V+
Sbjct: 7 FNSSPSKQVFLISTRAGGLGLNLVSANRVVIFDPNWNPAQDLQAQDRSFRFGQKRHVVVF 186
Query: 746 RLVVRASVEERIL--QLAKKKL 765
RL+ S+EE + Q+ K++L
Sbjct: 187 RLLAAGSLEELVYSRQVYKQQL 252
>TC228720 weakly similar to UP|RAD5_YEAST (P32849) DNA repair protein RAD5,
partial (4%)
Length = 736
Score = 71.6 bits (174), Expect = 1e-12
Identities = 41/103 (39%), Positives = 61/103 (58%)
Frame = +3
Query: 675 SVTDRQTAIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAH 734
SV R A+ F V ++S ++ LG+NL A V++ D +NP + QA++RAH
Sbjct: 3 SVVARDKAVKDFXNCPEVTVIIMSLKAASLGLNLVVACHVLMLDLWWNPTTEDQAIDRAH 182
Query: 735 RIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGS 777
RIGQ+ + V RL VR +VE+RIL L +KK M+ G+ G+
Sbjct: 183 RIGQTRPVTVLRLTVRDTVEDRILALQQKKRMMVASAFGEDGT 311
>AW100110 weakly similar to GP|18463957|gb chromatin complex subunit A101
{Zea mays}, partial (14%)
Length = 391
Score = 65.1 bits (157), Expect = 1e-10
Identities = 41/104 (39%), Positives = 59/104 (56%), Gaps = 2/104 (1%)
Frame = +3
Query: 575 GKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKIL 634
G+ + + N+ +QLRKVCNHP L+ DS L E+ + K LL +L+ L
Sbjct: 93 GRSVPAGMIRNLAIQLRKVCNHPDLLESAFDDSYLYPPLDEIGGQCG-KFHLLDRLLQRL 269
Query: 635 YKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYE--RVDGSVSV 676
+ H+VLIFSQ TK+LDI++ Y F K +E R+DG V +
Sbjct: 270 FARNHKVLIFSQWTKVLDIMDYY----FSEKGFEVCRIDGGVKL 389
>TC210166 similar to UP|Q7XJP0 (Q7XJP0) SNF2/SWI2 family global transcription
factor, partial (7%)
Length = 946
Score = 58.9 bits (141), Expect = 1e-08
Identities = 30/75 (40%), Positives = 47/75 (62%)
Frame = +1
Query: 690 KSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVV 749
KS V LL + G+NL A V++ + NP A+ QA++R HRIGQ N+ L++R +V
Sbjct: 70 KSIQVLLLLIQHGANGLNLLEAQHVVLVEPLLNPAAEAQAISRVHRIGQKNKTLIHRFIV 249
Query: 750 RASVEERILQLAKKK 764
+ +VEE I +L + +
Sbjct: 250 KDTVEESIYKLNRSR 294
>TC232388
Length = 664
Score = 58.5 bits (140), Expect = 1e-08
Identities = 33/90 (36%), Positives = 48/90 (52%)
Frame = +1
Query: 682 AIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNR 741
++ +FN VF S ++CG GI+L A +II D NP QA+ RA R GQ +
Sbjct: 10 SMEKFNNSPDARVFFGSIKACGEGISLVGASRIIILDVHLNPSVTRQAIGRAFRPGQMKK 189
Query: 742 LLVYRLVVRASVEERILQLAKKKLMLDQLF 771
+ VYRLV S EE KK ++ +++
Sbjct: 190 VFVYRLVSADSPEEEDHNTCFKKELISKMW 279
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,490,486
Number of Sequences: 63676
Number of extensions: 506231
Number of successful extensions: 1984
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1950
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1966
length of query: 935
length of database: 12,639,632
effective HSP length: 106
effective length of query: 829
effective length of database: 5,889,976
effective search space: 4882790104
effective search space used: 4882790104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146651.1


