

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.4 + phase: 0
(100 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
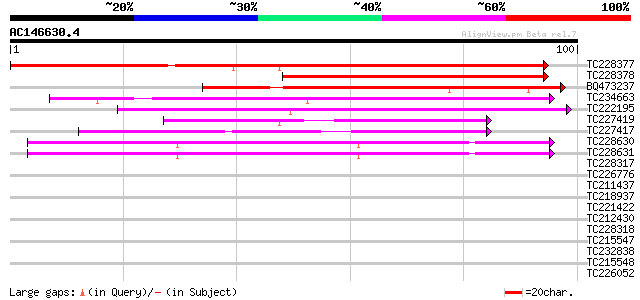
Score E
Sequences producing significant alignments: (bits) Value
TC228377 145 2e-36
TC228378 86 3e-18
BQ473237 77 2e-15
TC234663 75 7e-15
TC222195 72 6e-14
TC227419 52 5e-08
TC227417 51 8e-08
TC228630 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (64%) 40 1e-04
TC228631 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (50%) 40 1e-04
TC228317 34 0.013
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 34 0.013
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 33 0.017
TC218937 similar to UP|PEXA_ARATH (Q9SYU4) Peroxisome assembly p... 33 0.017
TC221422 33 0.022
TC212430 33 0.029
TC228318 32 0.038
TC215547 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partia... 31 0.085
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 31 0.11
TC215548 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partia... 30 0.15
TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_... 30 0.15
>TC228377
Length = 958
Score = 145 bits (367), Expect = 2e-36
Identities = 78/104 (75%), Positives = 86/104 (82%), Gaps = 9/104 (8%)
Frame = +1
Query: 1 MSRRNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSP---PSSCVSSE------NGSSS 51
MSRRN SGPKLDLKLNLSPPR +RR+ES PTRSA+ SP PSSCVSSE + S+S
Sbjct: 205 MSRRNESGPKLDLKLNLSPPRADRRLES-PTRSATASPTSPPSSCVSSELNQEDKSYSNS 381
Query: 52 PEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENN 95
PEATSM+LVGCPRCLMYVMLSE+DPKCPKC STVLLDFL+ N
Sbjct: 382 PEATSMVLVGCPRCLMYVMLSEDDPKCPKCKSTVLLDFLHDTKN 513
>TC228378
Length = 602
Score = 85.9 bits (211), Expect = 3e-18
Identities = 39/47 (82%), Positives = 43/47 (90%)
Frame = +3
Query: 49 SSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENN 95
S+SPEATSM+LVGCPRCLMYVMLSE+DPKCPKC STVLLDFL+ N
Sbjct: 12 SNSPEATSMVLVGCPRCLMYVMLSEDDPKCPKCKSTVLLDFLHDTKN 152
>BQ473237
Length = 408
Score = 76.6 bits (187), Expect = 2e-15
Identities = 42/68 (61%), Positives = 49/68 (71%), Gaps = 4/68 (5%)
Frame = -2
Query: 35 SVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSENDP--KCPKCHSTVLLDFL-- 90
S S P SCVSSE ++ E SM+LVGC RCLMYV+ S++DP KCPKC STVLLDFL
Sbjct: 386 SSSDPDSCVSSE--TNDEEIRSMVLVGCLRCLMYVLFSKDDPNPKCPKCKSTVLLDFLND 213
Query: 91 NAENNNNK 98
N + N NK
Sbjct: 212 NKQKNTNK 189
>TC234663
Length = 773
Score = 74.7 bits (182), Expect = 7e-15
Identities = 48/108 (44%), Positives = 56/108 (51%), Gaps = 19/108 (17%)
Frame = +3
Query: 8 GPKLDLK---LNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSS------------- 51
GP +L+ + SPP V +ESS SSC+SS SSS
Sbjct: 165 GPSSELRQPNMPTSPPEV---VESSSGELPEEFSWSSCLSSSVSSSSKIPIEDEENVQRA 335
Query: 52 ---PEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNN 96
PE +M LVGCPRC MYVMLSE DPKCPKC +TV L+ EN N
Sbjct: 336 NASPETKAMDLVGCPRCFMYVMLSEVDPKCPKCKTTVFLELFKDENFN 479
>TC222195
Length = 844
Score = 71.6 bits (174), Expect = 6e-14
Identities = 37/81 (45%), Positives = 47/81 (57%), Gaps = 1/81 (1%)
Frame = +1
Query: 20 PRVNRRMESSPTRSASVSPPSSCVSSENG-SSSPEATSMLLVGCPRCLMYVMLSENDPKC 78
P ++ +S S P S +E +S+PE +M LVGCPRC MYVMLSE DPKC
Sbjct: 232 PELSSSSSASSVSCTSKMPKESEEENEQRENSTPETKAMDLVGCPRCFMYVMLSEVDPKC 411
Query: 79 PKCHSTVLLDFLNAENNNNKK 99
PKC +TV L+ EN K+
Sbjct: 412 PKCKTTVFLELFKDENLKTKQ 474
>TC227419
Length = 688
Score = 52.0 bits (123), Expect = 5e-08
Identities = 29/64 (45%), Positives = 37/64 (57%), Gaps = 6/64 (9%)
Frame = +2
Query: 28 SSPTRSASVSPPSSCVSSE------NGSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
SSP+ S+S S + E N SSSP L GCP CL YVM+ +N+PKCP+C
Sbjct: 260 SSPSYSSSSSSIRETIQEEECEERLNVSSSP-----LAAGCPGCLSYVMIMKNNPKCPRC 424
Query: 82 HSTV 85
+S V
Sbjct: 425 NSLV 436
>TC227417
Length = 1067
Score = 51.2 bits (121), Expect = 8e-08
Identities = 30/73 (41%), Positives = 38/73 (51%)
Frame = +2
Query: 13 LKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLS 72
LK LS + SS R ++ C N SSSP A GCP CL YVM+
Sbjct: 596 LKSPLSVSSPSYSSSSSSIRETTIQE-DECEERLNVSSSPMAA-----GCPGCLSYVMIM 757
Query: 73 ENDPKCPKCHSTV 85
+N+PKCP+C+S V
Sbjct: 758 KNNPKCPRCNSLV 796
>TC228630 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (64%)
Length = 782
Score = 40.4 bits (93), Expect = 1e-04
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 12/105 (11%)
Frame = +3
Query: 4 RNGSGPKLDLKLNLSPPRVNRRMES-----------SPTRSASVSPPSSCVSSENGSSSP 52
RNG K D + + V+R ES S S+ P SS + G
Sbjct: 387 RNGMKAKEDPRSRRADYSVSRDCESEEEEEEESWYDSEESSSESCPSSSKEHYDEGQVEK 566
Query: 53 EATSMLLV-GCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNN 96
+ ++L+V GC CLMY M+ + CPKC + LL F +EN +
Sbjct: 567 QNNNVLVVAGCKGCLMYFMVPKQVEDCPKC-TGQLLHFDRSENGS 698
>TC228631 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (50%)
Length = 632
Score = 40.4 bits (93), Expect = 1e-04
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 12/105 (11%)
Frame = +1
Query: 4 RNGSGPKLDLKLNLSPPRVNRRMES-----------SPTRSASVSPPSSCVSSENGSSSP 52
RNG K D + + V+R ES S S+ P SS + G
Sbjct: 160 RNGMKAKEDPRSRRADYSVSRDCESEEEEEEESWYDSEESSSESCPSSSKEHYDEGQVEK 339
Query: 53 EATSMLLV-GCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNN 96
+ ++L+V GC CLMY M+ + CPKC + LL F +EN +
Sbjct: 340 QNNNVLVVAGCKGCLMYFMVPKQVEDCPKC-TGQLLHFDRSENGS 471
>TC228317
Length = 967
Score = 33.9 bits (76), Expect = 0.013
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +1
Query: 49 SSSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
SS + M+ C RC M VML ++ P CP C
Sbjct: 532 SSEDDYKEMVATVCMRCHMLVMLCKSSPSCPNC 630
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 33.9 bits (76), Expect = 0.013
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Frame = +2
Query: 2 SRRNGSGPKLDLKLNLSPPRVN---RRMESSPTRSASVSPPSSCVSSENGSSSPEATSM 57
S S PK + +PP +N R + P SA+ SPP S S N S++PE TS+
Sbjct: 782 SSSTSSSPKPTSNPSSNPPSLN*TSRSITLIPRSSAT*SPPPSDKPSNNPSTTPEITSL 958
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 33.5 bits (75), Expect = 0.017
Identities = 17/28 (60%), Positives = 20/28 (70%)
Frame = -1
Query: 29 SPTRSASVSPPSSCVSSENGSSSPEATS 56
SP S+S SPPSS S EN SSSP ++S
Sbjct: 497 SPLASSSGSPPSSPSSPENSSSSPSSSS 414
>TC218937 similar to UP|PEXA_ARATH (Q9SYU4) Peroxisome assembly protein 10
(Peroxin-10) (AthPEX10) (Pex10p) (PER8), partial (73%)
Length = 1217
Score = 33.5 bits (75), Expect = 0.017
Identities = 26/87 (29%), Positives = 39/87 (43%), Gaps = 4/87 (4%)
Frame = +1
Query: 3 RRNGSG-PKLDLKLNLSPPRVNRRM---ESSPTRSASVSPPSSCVSSENGSSSPEATSML 58
R G G P L+ + NL+ P +++R ESS + + S S C + P ATS
Sbjct: 829 RSAGHGLPVLNEEGNLATPDIDKRSWVSESSSSEYHATSGVSKCTLCLSNRQHPTATSCG 1008
Query: 59 LVGCPRCLMYVMLSENDPKCPKCHSTV 85
V C C+ P+CP C + +
Sbjct: 1009HVFCWNCI--TEWCNEKPECPLCRTPI 1083
>TC221422
Length = 1108
Score = 33.1 bits (74), Expect = 0.022
Identities = 17/44 (38%), Positives = 21/44 (47%)
Frame = +1
Query: 38 PPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
P S +SSE M+ C RC M VML ++ P CP C
Sbjct: 481 PAPSWLSSEGDDHK----EMIATVCMRCHMLVMLCKSSPACPNC 600
>TC212430
Length = 361
Score = 32.7 bits (73), Expect = 0.029
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Frame = +1
Query: 41 SCVSSEN-----GSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
SCV + + S + M+ C RC M VML ++ P CP C
Sbjct: 25 SCVLTRSPPWWLSSEGDDHKEMITTVCMRCHMLVMLCKSSPACPNC 162
>TC228318
Length = 651
Score = 32.3 bits (72), Expect = 0.038
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +1
Query: 50 SSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
S + M+ C RC M VML ++ P CP C
Sbjct: 61 SEDDYKEMVATVCMRCHMLVMLCKSSPSCPNC 156
>TC215547 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 797
Score = 31.2 bits (69), Expect = 0.085
Identities = 16/36 (44%), Positives = 19/36 (52%)
Frame = +2
Query: 28 SSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCP 63
SSPT + S SPPSS S +S P A S + P
Sbjct: 377 SSPTSARSSSPPSSTSGSVKSTSDPTARSATRISSP 484
Score = 26.9 bits (58), Expect = 1.6
Identities = 14/38 (36%), Positives = 24/38 (62%)
Frame = +2
Query: 15 LNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSP 52
LN +P N ++P+RS++ +PP+S S + +SSP
Sbjct: 281 LNPNPSIAN---SATPSRSSTRTPPASSPSPSSATSSP 385
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 30.8 bits (68), Expect = 0.11
Identities = 18/39 (46%), Positives = 23/39 (58%)
Frame = -3
Query: 18 SPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATS 56
SPPR SS + S+S SPPS SS + SSP ++S
Sbjct: 474 SPPRWTSPSSSSSSSSSS-SPPSGASSSSSSLSSPSSSS 361
Score = 25.8 bits (55), Expect = 3.6
Identities = 16/39 (41%), Positives = 22/39 (56%)
Frame = -3
Query: 18 SPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATS 56
SPP + + S+S+S PSS SS + SSS A+S
Sbjct: 420 SPP------SGASSSSSSLSSPSSSSSSSSSSSSSSASS 322
>TC215548 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 771
Score = 30.4 bits (67), Expect = 0.15
Identities = 16/36 (44%), Positives = 19/36 (52%)
Frame = +3
Query: 28 SSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCP 63
SSPT + S SPPSS S +S P A S + P
Sbjct: 390 SSPTSARSSSPPSSTSGSVKSTSVPTARSATRISSP 497
Score = 25.8 bits (55), Expect = 3.6
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = +3
Query: 28 SSPTRSASVSPPSSCVSSENGSSSP 52
++P+RS++ +PP+S S + +SSP
Sbjct: 324 ATPSRSSTRTPPASSPSPSSATSSP 398
>TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_13), partial
(39%)
Length = 941
Score = 30.4 bits (67), Expect = 0.15
Identities = 24/57 (42%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Frame = +1
Query: 6 GSGPKLDLKLNLSPPRVN--RRMESSPTRSASVS-----PPSSCVSSENGSSSPEAT 55
GS P+ +SPP R +SP+ S+SVS PP C SS G SSP AT
Sbjct: 385 GSPPR-----TISPPSATAPRTSSASPSPSSSVSAAAPSPPPPCTSS--GPSSPRAT 534
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,384,840
Number of Sequences: 63676
Number of extensions: 82560
Number of successful extensions: 1017
Number of sequences better than 10.0: 207
Number of HSP's better than 10.0 without gapping: 948
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1003
length of query: 100
length of database: 12,639,632
effective HSP length: 76
effective length of query: 24
effective length of database: 7,800,256
effective search space: 187206144
effective search space used: 187206144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146630.4


