

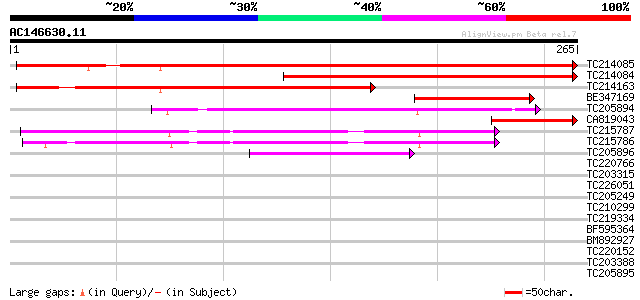
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.11 + phase: 0
(265 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214085 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, part... 424 e-119
TC214084 homologue to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor... 266 9e-72
TC214163 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, ... 244 4e-65
BE347169 114 3e-26
TC205894 similar to UP|Q8S3K4 (Q8S3K4) 1-cys peroxiredoxin, part... 89 2e-18
CA819043 homologue to GP|11558242|emb peroxiredoxin {Phaseolus v... 86 1e-17
TC215787 similar to UP|Q6QPJ6 (Q6QPJ6) Peroxiredoxin Q, complete 75 3e-14
TC215786 similar to UP|Q6QPJ6 (Q6QPJ6) Peroxiredoxin Q, partial ... 67 8e-12
TC205896 similar to UP|Q9SP12 (Q9SP12) 1-Cys peroxiredoxin, part... 52 4e-07
TC220766 homologue to UP|Q9Q5L3 (Q9Q5L3) EBNA-2, partial (4%) 33 0.17
TC203315 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, complete 33 0.17
TC226051 similar to UP|O24078 (O24078) Protein phosphatase 2C, p... 31 0.65
TC205249 similar to UP|Q8LBV7 (Q8LBV7) Contains similarity to pl... 30 0.86
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 30 1.1
TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting p... 30 1.1
BF595364 30 1.5
BM892927 30 1.5
TC220152 similar to UP|O65426 (O65426) Lsd1 like protein (At4g21... 30 1.5
TC203388 homologue to UP|H2B_GOSHI (O22582) Histone H2B, complete 29 1.9
TC205895 similar to UP|Q9SP12 (Q9SP12) 1-Cys peroxiredoxin, part... 29 2.5
>TC214085 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, partial (84%)
Length = 1129
Score = 424 bits (1091), Expect = e-119
Identities = 221/264 (83%), Positives = 233/264 (87%), Gaps = 2/264 (0%)
Frame = +1
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSI-PNASNSLPKLRTSLPLSLNRFTSSRRS 62
CSA SASL S+NP LFSPK S HL+ L P+ S + P L SL R + SRRS
Sbjct: 172 CSATSASLFSANPTPLFSPKPSLSLHLNPLPTRPSPSLTRPSL------SLTRPSHSRRS 333
Query: 63 FVVRASS-ELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTE 121
FVV+ASS ELPLVGN APDFEAEAVFDQEFI VKLS+YIGKKYV+LFFYPLDFTFVCPTE
Sbjct: 334 FVVKASSSELPLVGNTAPDFEAEAVFDQEFINVKLSDYIGKKYVVLFFYPLDFTFVCPTE 513
Query: 122 ITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKS 181
ITAFSDRHAEF LNTEILGVSVDSVFSHLAW+QTDRKSGGLGDLNYPL+SDVTKSISKS
Sbjct: 514 ITAFSDRHAEFEALNTEILGVSVDSVFSHLAWIQTDRKSGGLGDLNYPLISDVTKSISKS 693
Query: 182 YGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCP 241
YGVLIPDQGIALRGLFIIDKEGVIQHSTINNL IGRSVDETKRTLQALQYVQENPDEVCP
Sbjct: 694 YGVLIPDQGIALRGLFIIDKEGVIQHSTINNLAIGRSVDETKRTLQALQYVQENPDEVCP 873
Query: 242 AGWKPGEKSMKPDPKLSKEYFSAV 265
AGWKPGEKSMKPDPKLSK+YF+AV
Sbjct: 874 AGWKPGEKSMKPDPKLSKDYFAAV 945
>TC214084 homologue to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, partial
(52%)
Length = 1017
Score = 266 bits (679), Expect = 9e-72
Identities = 129/137 (94%), Positives = 134/137 (97%)
Frame = +2
Query: 129 HAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPD 188
HAEF LNTEILGVSVDSVFSHLAW+QTDRKSGGLGDLNYPL+SDVTKSISKSYGVLIPD
Sbjct: 416 HAEFEALNTEILGVSVDSVFSHLAWIQTDRKSGGLGDLNYPLISDVTKSISKSYGVLIPD 595
Query: 189 QGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGE 248
QGIALRGLFIIDKEGVI+HSTINNL IGRSVDETKRTLQALQYVQENPDEVCPAGWKPGE
Sbjct: 596 QGIALRGLFIIDKEGVIKHSTINNLAIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGE 775
Query: 249 KSMKPDPKLSKEYFSAV 265
KSMKPDPKLSK+YF+AV
Sbjct: 776 KSMKPDPKLSKDYFAAV 826
>TC214163 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, partial
(48%)
Length = 548
Score = 244 bits (622), Expect = 4e-65
Identities = 130/169 (76%), Positives = 139/169 (81%), Gaps = 1/169 (0%)
Frame = +2
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSF 63
CSA SASL S+NP LFSPK SSLS+PN S L L T LSL R + +RRSF
Sbjct: 62 CSATSASLFSANPTPLFSPK-------SSLSLPNNSLHLNPLPTRPSLSLTRPSHTRRSF 220
Query: 64 VVRASS-ELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEI 122
VV+ASS ELPLVGN APDFEAEAVFDQEFI VKLS+YIGKKYV+LFFYPLDFTFVCPTEI
Sbjct: 221 VVKASSSELPLVGNTAPDFEAEAVFDQEFINVKLSDYIGKKYVVLFFYPLDFTFVCPTEI 400
Query: 123 TAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLV 171
TAFSDRHAEF LNTEIL VS DSVFSHLAW+QTDRKSGGLGDLNYPL+
Sbjct: 401 TAFSDRHAEFEALNTEILSVSDDSVFSHLAWIQTDRKSGGLGDLNYPLI 547
>BE347169
Length = 471
Score = 114 bits (286), Expect = 3e-26
Identities = 55/56 (98%), Positives = 55/56 (98%)
Frame = +3
Query: 190 GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWK 245
GIALRGLFIIDKEGVIQHSTINNL IGRSVDETKRTLQALQYVQENPDEVCPAGWK
Sbjct: 303 GIALRGLFIIDKEGVIQHSTINNLAIGRSVDETKRTLQALQYVQENPDEVCPAGWK 470
>TC205894 similar to UP|Q8S3K4 (Q8S3K4) 1-cys peroxiredoxin, partial (96%)
Length = 854
Score = 89.4 bits (220), Expect = 2e-18
Identities = 56/189 (29%), Positives = 96/189 (50%), Gaps = 7/189 (3%)
Frame = +2
Query: 67 ASSELP--LVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITA 124
++S++P +G+ PD + E K+KL ++ + ILF +P DFT VC TE+
Sbjct: 8 SASKMPGLTIGDTIPDLQVETNQG----KIKLHQFCADGWTILFSHPGDFTPVCTTELGK 175
Query: 125 FSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGV 184
+ EF + ++LG+S D V SH W++ +NYP+++D + I K +
Sbjct: 176 MAQYAKEFYQRGVKLLGLSCDDVQSHNEWIKDIEAYTPGAKVNYPIIADPKREIIKQLNM 355
Query: 185 LIPDQ-----GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEV 239
+ PD+ + R L I+ + I+ S + GR++DE R +++LQ +
Sbjct: 356 VDPDEKDSTGNLPSRALHIVGPDLKIKLSFLYPATTGRNMDEVLRVIESLQKASKF-KVA 532
Query: 240 CPAGWKPGE 248
PA WKPG+
Sbjct: 533 TPANWKPGD 559
>CA819043 homologue to GP|11558242|emb peroxiredoxin {Phaseolus vulgaris},
partial (17%)
Length = 444
Score = 86.3 bits (212), Expect = 1e-17
Identities = 38/40 (95%), Positives = 40/40 (100%)
Frame = +3
Query: 226 LQALQYVQENPDEVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
LQALQYVQENPDEVCPAGWKPGEKSMKPDPKLSK+YF+AV
Sbjct: 159 LQALQYVQENPDEVCPAGWKPGEKSMKPDPKLSKDYFAAV 278
>TC215787 similar to UP|Q6QPJ6 (Q6QPJ6) Peroxiredoxin Q, complete
Length = 929
Score = 75.1 bits (183), Expect = 3e-14
Identities = 67/226 (29%), Positives = 105/226 (45%), Gaps = 2/226 (0%)
Frame = +3
Query: 6 APSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVV 65
+P ASL N + + SP H SS ++P S L LS + +S S +
Sbjct: 24 SPMASLTVPNHCVPTLLRTHSPNHPSSQNLPFPSTPSNSQFFGLKLSHSSVSSIPSSSSL 203
Query: 66 RASSELPL-VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITA 124
+ + + G+ P+F + DQ V LS + GK VI++FYP D T C + A
Sbjct: 204 KGTIFAKVNKGSKPPNFTLK---DQNGKNVSLSNFKGKP-VIVYFYPADETPGCTKQACA 371
Query: 125 FSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGV 184
F D + +F + ++G+S D SH A+ R L + L+SD + K +GV
Sbjct: 372 FRDSYEKFKKAGAVVVGISGDDAASHKAFASKYR-------LPFTLLSDEGNKVRKEWGV 530
Query: 185 LIPDQG-IALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQAL 229
G + R +++DK GV+Q N + +DET + LQ+L
Sbjct: 531 PGDFFGSLPGRETYVLDKNGVVQLVYNNQFQPEKHIDETLKILQSL 668
>TC215786 similar to UP|Q6QPJ6 (Q6QPJ6) Peroxiredoxin Q, partial (89%)
Length = 1085
Score = 67.0 bits (162), Expect = 8e-12
Identities = 67/229 (29%), Positives = 104/229 (45%), Gaps = 6/229 (2%)
Frame = +2
Query: 7 PSASLLSSN---PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSF 63
P ASL N P +L + L+ P SS ++P S L LS + F++ S
Sbjct: 143 PMASLTVPNHCVPTLLRTHSLNYP---SSQNLPLPSAPSNSQFFGLKLSHSSFSTIPSSS 313
Query: 64 VVRASSELPLV--GNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTE 121
S V G+ P+F + DQ V LS + GK V+++FYP D T C +
Sbjct: 314 SSLKGSIFAKVTKGSKPPNFTLK---DQNGKNVSLSNFKGKP-VVVYFYPADETPGCTKQ 481
Query: 122 ITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKS 181
AF D + +F + ++G+S D SH A+ + L + L+SD + K
Sbjct: 482 ACAFRDSYEKFKKAGAVVVGISGDDAASHKAFASKYK-------LPFTLLSDEGNKVRKE 640
Query: 182 YGVLIPDQG-IALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQAL 229
+GV G + R +++DK GV+Q N + + ET + LQ+L
Sbjct: 641 WGVPGDFFGSLPGRETYVLDKNGVVQLVYNNQFQPEKHIGETLKILQSL 787
>TC205896 similar to UP|Q9SP12 (Q9SP12) 1-Cys peroxiredoxin, partial (75%)
Length = 714
Score = 51.6 bits (122), Expect = 4e-07
Identities = 25/77 (32%), Positives = 41/77 (52%)
Frame = +1
Query: 113 DFTFVCPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVS 172
DFT VC TE+ + EF + ++LG+S D V SH W++ +NYP+++
Sbjct: 232 DFTPVCTTELGKMAQYAKEFYQRGVKLLGLSCDDVQSHNEWIKDIEAYTPGAKVNYPIIA 411
Query: 173 DVTKSISKSYGVLIPDQ 189
D + I K ++ PD+
Sbjct: 412 DPKREIIKQLNMVDPDE 462
>TC220766 homologue to UP|Q9Q5L3 (Q9Q5L3) EBNA-2, partial (4%)
Length = 885
Score = 32.7 bits (73), Expect = 0.17
Identities = 27/78 (34%), Positives = 35/78 (44%), Gaps = 3/78 (3%)
Frame = +3
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSL---NRFTSSRR 61
S+PS + LSSNP S S PP + LP + T LPLSL N SS+
Sbjct: 6 SSPSPTTLSSNPKPSLSSTASDPPSAKPY------HPLPTIITILPLSLSNPNSPPSSKT 167
Query: 62 SFVVRASSELPLVGNAAP 79
F ++ L A+P
Sbjct: 168RFTTTTTTTLHHQLQASP 221
>TC203315 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, complete
Length = 778
Score = 32.7 bits (73), Expect = 0.17
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 2/64 (3%)
Frame = -2
Query: 2 ACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPN--ASNSLPKLRTSLPLSLNRFTSS 59
+C S S+S SS+ L + SSPP SS +P+ ++S P LP L + +S
Sbjct: 246 SCRSPRSSSCASSSSRSLLPSRSSSPPSASSPPVPSPPLASSPPASSLPLPEPLGKSETS 67
Query: 60 RRSF 63
++ F
Sbjct: 66 KKGF 55
>TC226051 similar to UP|O24078 (O24078) Protein phosphatase 2C, partial
(86%)
Length = 1450
Score = 30.8 bits (68), Expect = 0.65
Identities = 17/41 (41%), Positives = 26/41 (62%), Gaps = 1/41 (2%)
Frame = +1
Query: 4 CSAPSASLLSSNPNILFSPKLSSPP-HLSSLSIPNASNSLP 43
CS+PSA+ SS+P+ F +L PP S+ S +AS++ P
Sbjct: 16 CSSPSAAAASSSPSSPFRLRLPKPPTAFSASSAASASSTSP 138
>TC205249 similar to UP|Q8LBV7 (Q8LBV7) Contains similarity to plastid
ribosomal protein L19, partial (54%)
Length = 962
Score = 30.4 bits (67), Expect = 0.86
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 17/100 (17%)
Frame = +1
Query: 19 LFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELPLVGNAA 78
LF+PKLS P ++++ + S P PL ++ SFVVRA + L G A
Sbjct: 139 LFAPKLSFPSSSRNVALISRVPSTPISWRCTPLI------AKPSFVVRADTNLDGGGEAT 300
Query: 79 PDF-----------EAEAVFDQEFIK------VKLSEYIG 101
D EAE V D E K VKL + +G
Sbjct: 301 EDVPSDNAEKVSEGEAEQVSDSEAPKSPRKPRVKLGDIMG 420
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 30.0 bits (66), Expect = 1.1
Identities = 24/67 (35%), Positives = 34/67 (49%), Gaps = 3/67 (4%)
Frame = +1
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNA---SNSLPKLRTSLPLSLNRFTSSRR 61
+A + S L+ P++ FSP SS P+A S+S T L L+ T+ RR
Sbjct: 16 AAATTSKLAYPPHVHFSPSPSSNYLFLKTHKPSATHLSSSFIHPTTILHLAAANTTTRRR 195
Query: 62 SFVVRAS 68
SF VRA+
Sbjct: 196SFTVRAA 216
>TC219334 weakly similar to UP|Q9FUZ9 (Q9FUZ9) SKP1 interacting partner 6
(Fragment), partial (29%)
Length = 1451
Score = 30.0 bits (66), Expect = 1.1
Identities = 20/60 (33%), Positives = 30/60 (49%)
Frame = +1
Query: 13 SSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELP 72
S NP+ S SS PH S + PN S++ T+ PLS N S+ ++ + +S P
Sbjct: 142 SPNPSTPSSLPQSSSPHEPSSNAPNPSSTSLSTHTT-PLSSNSSRSTTQTLITPSSLPFP 318
>BF595364
Length = 418
Score = 29.6 bits (65), Expect = 1.5
Identities = 13/39 (33%), Positives = 21/39 (53%)
Frame = +2
Query: 12 LSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLP 50
L PN+ F P+ PP + +P+ + L +LR +LP
Sbjct: 77 LQEAPNLPFVPRHPQPPFQHQIPLPSHRHHLRRLRRALP 193
>BM892927
Length = 428
Score = 29.6 bits (65), Expect = 1.5
Identities = 18/38 (47%), Positives = 23/38 (60%)
Frame = +3
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSL 42
S P +SL+ S +P LSSPP S SIP+AS S+
Sbjct: 87 STPGSSLIPSPRFCTPTPNLSSPPS-SKKSIPSASTSM 197
>TC220152 similar to UP|O65426 (O65426) Lsd1 like protein (At4g21610),
partial (61%)
Length = 701
Score = 29.6 bits (65), Expect = 1.5
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +2
Query: 224 RTLQALQYVQENPDEVCPAGWKPGEKSMKPDPKLS 258
R +Q+ Q V+E DE P GW+P P P+ S
Sbjct: 122 RKMQSQQTVKEEDDEGPPPGWQPLPAQPPPPPRPS 226
>TC203388 homologue to UP|H2B_GOSHI (O22582) Histone H2B, complete
Length = 675
Score = 29.3 bits (64), Expect = 1.9
Identities = 20/49 (40%), Positives = 24/49 (48%)
Frame = -2
Query: 11 LLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSS 59
L SS P P +SSPP SSL L +SLPL +RF +S
Sbjct: 179 LASSPPEPCLPPSISSPPRASSL-----------LASSLPLVPSRFGAS 66
>TC205895 similar to UP|Q9SP12 (Q9SP12) 1-Cys peroxiredoxin, partial (47%)
Length = 432
Score = 28.9 bits (63), Expect = 2.5
Identities = 13/33 (39%), Positives = 20/33 (60%)
Frame = +2
Query: 216 GRSVDETKRTLQALQYVQENPDEVCPAGWKPGE 248
GR++DE R +++LQ + PA WKPG+
Sbjct: 206 GRNMDEVLRVIESLQKASKFK-VATPANWKPGD 301
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,783,467
Number of Sequences: 63676
Number of extensions: 163993
Number of successful extensions: 1343
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1321
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1332
length of query: 265
length of database: 12,639,632
effective HSP length: 96
effective length of query: 169
effective length of database: 6,526,736
effective search space: 1103018384
effective search space used: 1103018384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146630.11


