

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
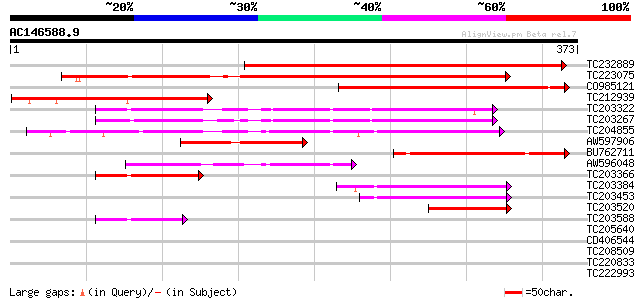
Query= AC146588.9 + phase: 0
(373 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232889 weakly similar to UP|O65340 (O65340) Metalloproteinase ... 341 4e-94
TC223075 UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1 precurso... 301 4e-82
CO985121 197 5e-51
TC212939 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, p... 165 4e-41
TC203322 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 142 2e-34
TC203267 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 142 3e-34
TC204855 UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2, complete 136 2e-32
AW597906 weakly similar to PIR|T00643|T006 zinc metalloproteinas... 106 2e-23
BU762711 103 1e-22
AW596048 weakly similar to GP|16901508|gb| matrix metalloprotein... 93 2e-19
TC203366 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 67 1e-11
TC203384 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 67 2e-11
TC203453 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 60 2e-09
TC203520 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 52 3e-07
TC203588 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase ... 49 5e-06
TC205640 similar to UP|Q9FHW0 (Q9FHW0) Gb|AAF07835.1 (At5g37070)... 31 1.0
CD406544 30 1.8
TC208509 similar to UP|Q9LRP8 (Q9LRP8) Non-phototropic hypocotyl... 29 3.0
TC220833 similar to UP|Q9FL00 (Q9FL00) Emb|CAB62433.1|, partial ... 28 6.7
TC222993 28 6.7
>TC232889 weakly similar to UP|O65340 (O65340) Metalloproteinase , partial
(38%)
Length = 734
Score = 341 bits (874), Expect = 4e-94
Identities = 163/212 (76%), Positives = 185/212 (86%)
Frame = +1
Query: 155 VSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLKFTETTLYS 214
V+HFT+FPG PRWPEG QELTYAFFPGN L++ VK VFA AFARW+EVT+LKF ET Y
Sbjct: 1 VAHFTLFPGMPRWPEGTQELTYAFFPGNGLSDAVKGVFAAAFARWAEVTSLKFRETASYF 180
Query: 215 GADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAV 274
GADI+IGFF+GDHGDGEPFDGSLGTLAHAFSP NGRFHLDAAEDWVVSGDV++S+LPTAV
Sbjct: 181 GADIRIGFFSGDHGDGEPFDGSLGTLAHAFSPTNGRFHLDAAEDWVVSGDVTRSALPTAV 360
Query: 275 DLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGS 334
DLESVAVHEIGHLLGLGHSS EEA+MFPTISSR KKVVLA DD+ GIQ+LYG+NP+FNGS
Sbjct: 361 DLESVAVHEIGHLLGLGHSSVEEAVMFPTISSRKKKVVLARDDIEGIQFLYGSNPNFNGS 540
Query: 335 TVISSPERNIGNGGCSLTSLWSPWRLFSLLTF 366
T S+PER+ +GG LT + +F+LLTF
Sbjct: 541 TATSAPERDASDGGRCLTCVGPSSGVFALLTF 636
>TC223075 UP|MEP1_SOYBN (P29136) Metalloendoproteinase 1 precursor (SMEP1) ,
complete
Length = 1198
Score = 301 bits (770), Expect = 4e-82
Identities = 162/299 (54%), Positives = 194/299 (64%), Gaps = 4/299 (1%)
Frame = +3
Query: 35 PGTLPVGP--WD--AFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDL 90
P GP WD A FT G+NY GLSN+KNYF H GYIP +P +F D+FDD L
Sbjct: 114 PSVSAHGPYAWDGEATYKFTTYHPGQNYKGLSNVKNYFHHLGYIPNAP--HFDDNFDDTL 287
Query: 91 QEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKL 150
AIKTYQKN+NLNVTG+ D TL+Q+M PRCGV DII T +TTS
Sbjct: 288 VSAIKTYQKNYNLNVTGKFDINTLKQIMTPRCGVPDIIINTN--------KTTS------ 425
Query: 151 RFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLKFTET 210
F +S +T F PRW G +LTYAF P L +T KS A AF++W+ V + F ET
Sbjct: 426 -FGMISDYTFFKDMPRWQAGTTQLTYAFSPEPRLDDTFKSAIARAFSKWTPVVNIAFQET 602
Query: 211 TLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSL 270
T Y A+IKI F + +HGD PFDG G L HAF+P +GR H DA E WV SGDV+KS +
Sbjct: 603 TSYETANIKILFASKNHGDPYPFDGPGGILGHAFAPTDGRCHFDADEYWVASGDVTKSPV 782
Query: 271 PTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNP 329
+A DLESVAVHEIGHLLGLGHSS+ AIM+P+I R +KV LA DD+ GI+ LYG NP
Sbjct: 783 TSAFDLESVAVHEIGHLLGLGHSSDLRAIMYPSIPPRTRKVNLAQDDIDGIRKLYGINP 959
>CO985121
Length = 535
Score = 197 bits (502), Expect = 5e-51
Identities = 98/152 (64%), Positives = 118/152 (77%)
Frame = -2
Query: 217 DIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDL 276
DI+IGF+ GDHGDGEPFDG LGTLAHA P NG FHLD+AEDWV SGDV+K+SL AVDL
Sbjct: 534 DIRIGFYGGDHGDGEPFDGVLGTLAHAXXPTNGMFHLDSAEDWVASGDVTKASLSNAVDL 355
Query: 277 ESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTV 336
ESVAVHEIGHLLGLGHSS E+AIM+PTI++R +KV L +DD++GIQ LYG+NP+F G+
Sbjct: 354 ESVAVHEIGHLLGLGHSSVEDAIMYPTITARTRKVELNEDDIQGIQVLYGSNPNFTGTPA 175
Query: 337 ISSPERNIGNGGCSLTSLWSPWRLFSLLTFVL 368
SS E + +GG + FSLL FV+
Sbjct: 174 TSSRESDTSDGGVRHVGA-TCGVFFSLLLFVV 82
>TC212939 weakly similar to UP|Q9ZUJ5 (Q9ZUJ5) T2K10.2 protein, partial (13%)
Length = 821
Score = 165 bits (417), Expect = 4e-41
Identities = 83/140 (59%), Positives = 100/140 (71%), Gaps = 8/140 (5%)
Frame = +1
Query: 2 KKQQLIFPLT---ISFSLTLLIVSARLFPDV---PSWIPPGTLPVGPWDAFRNFTGCRQG 55
+++ L+F T + FS VSARLFP+V P W P W+A+RNFTGCR G
Sbjct: 400 QQRYLLFAFTTLIVFFSFNTTPVSARLFPNVSSIPRWNTSHAPPDSAWNAYRNFTGCRPG 579
Query: 56 ENYNGLSNLKNYFQHFGYIPR--SPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMT 113
+ YNGLSNLKNYF +FGYI S SNFSDDFDD L+ A++ YQKNFNLN+TGELDD T
Sbjct: 580 KTYNGLSNLKNYFHYFGYISNVSSKSSNFSDDFDDALEAAVRAYQKNFNLNITGELDDPT 759
Query: 114 LRQVMLPRCGVADIINGTTT 133
+ Q++ PRCGVADIINGTTT
Sbjct: 760 MNQIVKPRCGVADIINGTTT 819
>TC203322 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (74%)
Length = 1664
Score = 142 bits (359), Expect = 2e-34
Identities = 95/270 (35%), Positives = 142/270 (52%), Gaps = 5/270 (1%)
Frame = +2
Query: 57 NYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQ 116
N GLS +K+Y +GYI S F+D FD ++ AIKTYQ NL VTG L+ ++Q
Sbjct: 413 NIKGLSVVKDYLSDYGYIESS--GPFTDSFDQEIISAIKTYQNFSNLQVTGGLNKQLIQQ 586
Query: 117 VMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTY 176
++ RCGV D+ + +D + + H RW + LTY
Sbjct: 587 MLSIRCGVPDV------------NFDYNFTDDNISYPKAGH--------RWFPNRN-LTY 703
Query: 177 AFFPGNELTETVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHGDGEPFDG 235
F P N++ + + VF +FARW++ + TL ETT Y ADI++GF+N + E + G
Sbjct: 704 GFLPENQIPDNMTKVFRDSFARWAQASGTLSLRETT-YDNADIQVGFYNFTNSRIEVYGG 880
Query: 236 SLGTLAHAFSPRNGRFHLDAAEDWVV-SGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSS 294
S S + G LD W++ S + + S +DLE+VA+H+IGHLLGL HS
Sbjct: 881 ST-IFLQPDSSKKGVVLLDGNMGWLLPSENATLSKDDGVLDLETVAMHQIGHLLGLDHSH 1057
Query: 295 EEEAIMFPTI---SSRMKKVVLADDDVRGI 321
+E+++M+P I S +KV L++ D I
Sbjct: 1058KEDSVMYPYILSSQSEQRKVQLSNSDKANI 1147
>TC203267 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (61%)
Length = 1338
Score = 142 bits (357), Expect = 3e-34
Identities = 97/268 (36%), Positives = 145/268 (53%), Gaps = 3/268 (1%)
Frame = +3
Query: 57 NYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQ 116
N GLS +K+Y +GYI S F++ FD + AIKTYQK NL VTG + ++Q
Sbjct: 294 NIKGLSVVKDYLSEYGYIESSRP--FNNSFDQETMSAIKTYQKFSNLPVTGVPNKQLIQQ 467
Query: 117 VMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTY 176
++ RCGV D+ D T ++ S + H RW + LTY
Sbjct: 468 MLSLRCGVPDV---------NFDYNFTDDNTS---YPKAGH--------RWFPNRN-LTY 584
Query: 177 AFFPGNELTETVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHGDGEPFDG 235
F P N++ + + VF +FARW++ + TL TETT Y ADI++GF+N + E + G
Sbjct: 585 GFLPENQIPDNMTKVFRDSFARWAQASGTLSLTETT-YDNADIQVGFYNFTNSRIEVYGG 761
Query: 236 SLGTLAHAFSPRNGRFHLDAAEDWVV-SGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSS 294
SL S + G L+ W++ S + + S +DLE+ A+H+IGHLLGL HS
Sbjct: 762 SL-IFLQPDSSKKGVVLLNGNMGWLLPSENATLSKDDGVLDLETAAMHQIGHLLGLDHSH 938
Query: 295 EEEAIMFPTI-SSRMKKVVLADDDVRGI 321
+E+++M+P I SS+ +KV L++ D I
Sbjct: 939 KEDSVMYPYILSSQQRKVQLSNSDKANI 1022
>TC204855 UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2, complete
Length = 1307
Score = 136 bits (342), Expect = 2e-32
Identities = 111/325 (34%), Positives = 165/325 (50%), Gaps = 11/325 (3%)
Frame = +1
Query: 12 ISFSLTLLIVSARL--FPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNG--LSNLKNY 67
+SFS L + +L P + ++ P T +G NFT E + +S +K+Y
Sbjct: 124 VSFSSFLKQLKQKLEKSPTLKDFLKPTT--IGDIYYTLNFTEIFSSEERSAPPVSLIKDY 297
Query: 68 FQHFGYIPRS-PKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVAD 126
++GYI S P SN D + + AIKTYQ+ F+L TG+L++ TL+Q+ RCGV D
Sbjct: 298 LSNYGYIESSGPLSNSMDQ--ETIISAIKTYQQYFSLQPTGKLNNETLQQMSFLRCGVPD 471
Query: 127 IINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTE 186
I + +D + + H RW LTY F P N++
Sbjct: 472 I------------NIDYNFTDDNMSYPKAGH--------RWFP-HTNLTYGFLPENQIPA 588
Query: 187 TVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHG--DGEPFDGSLGTLAHA 243
+ VF +FARW++ + L TETT Y ADI++GF+N + D E + GSL L
Sbjct: 589 NMTKVFRDSFARWAQASGVLNLTETT-YDNADIQVGFYNFTYLGIDIEVYGGSLIFL-QP 762
Query: 244 FSPRNGRFHLDAAED-WVVSGDVSKSSLPTAV-DLESVAVHEIGHLLGLGHSSEEEAIMF 301
S + G LD W + + + S V DLES A+HEIGHLLGL HS++E+++M+
Sbjct: 763 DSTKKGVILLDGTNKLWALPSENGRLSWEEGVLDLESAAMHEIGHLLGLDHSNKEDSVMY 942
Query: 302 PTI-SSRMKKVVLADDDVRGIQYLY 325
P I S +KV L+ D +Q+ +
Sbjct: 943 PCILPSHQRKVQLSKSDKTNVQHQF 1017
>AW597906 weakly similar to PIR|T00643|T006 zinc metalloproteinase homolog
F3I6.6 - Arabidopsis thaliana, partial (11%)
Length = 279
Score = 106 bits (264), Expect = 2e-23
Identities = 50/84 (59%), Positives = 61/84 (72%)
Frame = -2
Query: 113 TLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQ 172
T+ Q++ PRCGVADIINGTTTMN+GK T S + FHTV+H++ F GQPRWP G Q
Sbjct: 239 TMNQIVKPRCGVADIINGTTTMNSGKTNTTDSPT-----FHTVAHYSFFDGQPRWPVGTQ 75
Query: 173 ELTYAFFPGNELTETVKSVFATAF 196
ELTYAF P N L + +K+VF AF
Sbjct: 74 ELTYAFDPDNALDDVIKTVFGNAF 3
>BU762711
Length = 428
Score = 103 bits (258), Expect = 1e-22
Identities = 59/116 (50%), Positives = 75/116 (63%)
Frame = +1
Query: 253 LDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVV 312
LDAAE W V D + AVDLESVA HEIGH+LGLGHSS +EA+M+P++S R KKV
Sbjct: 1 LDAAETWSV--DFEREESRVAVDLESVATHEIGHVLGLGHSSVKEAVMYPSLSPRRKKVD 174
Query: 313 LADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSLLTFVL 368
L DDV G+Q LYG+NP+F S+++ S +N N L + + W L L F L
Sbjct: 175 LRIDDVVGVQALYGSNPNFTFSSLLQS--QNSLNAAVGLETGFYKWTLTLPLAFFL 336
>AW596048 weakly similar to GP|16901508|gb| matrix metalloproteinase MMP2
{Glycine max}, partial (16%)
Length = 417
Score = 93.2 bits (230), Expect = 2e-19
Identities = 60/154 (38%), Positives = 79/154 (50%), Gaps = 2/154 (1%)
Frame = +3
Query: 77 SPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNA 136
S F+DD D AI TYQ+ FNL +TG+L + TL+Q+ LPRCGV D MN
Sbjct: 9 SSGGTFNDDLDQATVSAITTYQRFFNLKITGDLTNETLQQISLPRCGVPD-------MNF 167
Query: 137 GKDTETTSNSDSKLRFHTVSHFTVFPGQPRW-PEGKQELTYAFFPGNELTETVKSVFATA 195
D + S R+H RW P+ + LTY F P +++ VF A
Sbjct: 168 DYDVSKDNVSWPMSRYHR-----------RWFPD--RNLTYGFSPASKIPSNATKVFRDA 308
Query: 196 FARWS-EVTTLKFTETTLYSGADIKIGFFNGDHG 228
FARW+ V L TE Y+ AD+K+GF+N D G
Sbjct: 309 FARWAGSVPGLNLTEMN-YNSADLKVGFYNLDEG 407
>TC203366 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (18%)
Length = 576
Score = 67.4 bits (163), Expect = 1e-11
Identities = 33/71 (46%), Positives = 46/71 (64%)
Frame = +1
Query: 57 NYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQ 116
N GLS +K+Y +GYI S F+D FD ++ AIKTYQ NLNVTG+L+ ++Q
Sbjct: 328 NIKGLSVVKDYLSDYGYIESS--GPFNDSFDQEIISAIKTYQNFSNLNVTGDLNKQLIQQ 501
Query: 117 VMLPRCGVADI 127
++ RCGV D+
Sbjct: 502 ILSIRCGVPDV 534
>TC203384 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (32%)
Length = 643
Score = 66.6 bits (161), Expect = 2e-11
Identities = 44/119 (36%), Positives = 70/119 (57%), Gaps = 4/119 (3%)
Frame = +1
Query: 216 ADIKIGFFNGD--HGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTA 273
ADI++GF+N + E + GSL L S + G +D W++ + + S
Sbjct: 7 ADIQVGFYNFTALSIEVEVYGGSLIFL-QPDSSKKGMVLMDGNMGWLLPSENATLSKDDG 183
Query: 274 V-DLESVAVHEIGHLLGLGHSSEEEAIMFPTI-SSRMKKVVLADDDVRGIQYLYGTNPS 330
V DLE+VA+H+IGHLLGL HS +E+++M+P I SS+ +KV L++ D I + ++ S
Sbjct: 184 VLDLETVAMHQIGHLLGLDHSHKEDSVMYPYILSSQQRKVKLSNSDKANIHLQFASHDS 360
>TC203453 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (29%)
Length = 622
Score = 59.7 bits (143), Expect = 2e-09
Identities = 38/102 (37%), Positives = 60/102 (58%), Gaps = 2/102 (1%)
Frame = +1
Query: 231 EPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAV-DLESVAVHEIGHLLG 289
E + GSL L S + G +D W++ + + S V DLE+VA+H+IGHLLG
Sbjct: 31 EVYGGSLIFL-QPDSSKKGVVLMDGNMGWLLPSENATLSKDDGVLDLETVAMHQIGHLLG 207
Query: 290 LGHSSEEEAIMFPTI-SSRMKKVVLADDDVRGIQYLYGTNPS 330
L HS +E+++M+P I SS+ +KV L++ D I + ++ S
Sbjct: 208 LDHSHKEDSVMYPYILSSQQRKVKLSNSDKANIHLQFASHDS 333
>TC203520 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (13%)
Length = 434
Score = 52.4 bits (124), Expect = 3e-07
Identities = 26/56 (46%), Positives = 40/56 (71%), Gaps = 1/56 (1%)
Frame = +2
Query: 276 LESVAVHEIGHLLGLGHSSEEEAIMFPTI-SSRMKKVVLADDDVRGIQYLYGTNPS 330
LE+VA+H+IGHLLGL HS +E+++M+P I SS+ +KV L++ D I + + S
Sbjct: 2 LETVAMHQIGHLLGLEHSPKEDSVMYPYILSSQQRKVKLSNSDKANIHLEFAKHDS 169
>TC203588 similar to UP|Q93Z89 (Q93Z89) Matrix metalloproteinase MMP2,
partial (14%)
Length = 514
Score = 48.5 bits (114), Expect = 5e-06
Identities = 26/61 (42%), Positives = 36/61 (58%)
Frame = +3
Query: 57 NYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQ 116
N GL +K+Y +GYI S F+D FD ++ AIKTYQ NL VTG L+ ++Q
Sbjct: 336 NITGLYIVKDYLSDYGYIESS--GPFNDSFDQEIISAIKTYQNFSNLQVTGGLNKELIQQ 509
Query: 117 V 117
+
Sbjct: 510 M 512
>TC205640 similar to UP|Q9FHW0 (Q9FHW0) Gb|AAF07835.1 (At5g37070), complete
Length = 996
Score = 30.8 bits (68), Expect = 1.0
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Frame = -3
Query: 138 KDTETTSNSDSKLRFHTVSHFTVF-PGQPRWPEGKQELTY 176
K TETTS++ ++ FH VS FT F + +W + L Y
Sbjct: 730 KKTETTSSTRTQEEFHQVSGFTTFKSNRIKWSTRRDNLFY 611
>CD406544
Length = 484
Score = 30.0 bits (66), Expect = 1.8
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +2
Query: 3 KQQLIFPLTISFSLTLLIVSARLFPDV 29
K +I IS SLT ++VSARLFP+V
Sbjct: 404 KPIIILAFLISLSLTNVVVSARLFPNV 484
>TC208509 similar to UP|Q9LRP8 (Q9LRP8) Non-phototropic hypocotyl-like
protein, partial (63%)
Length = 972
Score = 29.3 bits (64), Expect = 3.0
Identities = 15/37 (40%), Positives = 20/37 (53%)
Frame = +1
Query: 334 STVISSPERNIGNGGCSLTSLWSPWRLFSLLTFVLSH 370
S + S+P + G SLW PW +F LL +LSH
Sbjct: 163 SPLTSNPPKASQIHG*RRGSLWKPW*VFCLLRRILSH 273
>TC220833 similar to UP|Q9FL00 (Q9FL00) Emb|CAB62433.1|, partial (32%)
Length = 626
Score = 28.1 bits (61), Expect = 6.7
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 5/49 (10%)
Frame = +1
Query: 56 ENYNGLSNLKN-----YFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQK 99
E YN + KN + H P SP+ +FS+DF D+Q+A+K ++
Sbjct: 100 EMYNNSEHNKNNKGDNHHHHHQCAPMSPRISFSNDF-VDVQQAMKLQEQ 243
>TC222993
Length = 855
Score = 28.1 bits (61), Expect = 6.7
Identities = 13/22 (59%), Positives = 15/22 (68%)
Frame = -1
Query: 14 FSLTLLIVSARLFPDVPSWIPP 35
F+L L SARLFPD + IPP
Sbjct: 717 FTLFLFFFSARLFPDPVATIPP 652
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,914,970
Number of Sequences: 63676
Number of extensions: 232787
Number of successful extensions: 1287
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1264
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1269
length of query: 373
length of database: 12,639,632
effective HSP length: 99
effective length of query: 274
effective length of database: 6,335,708
effective search space: 1735983992
effective search space used: 1735983992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146588.9


