

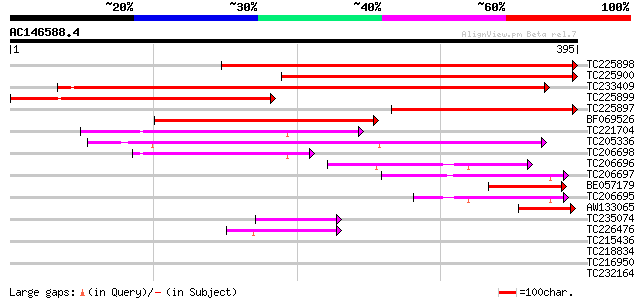
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
(395 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225898 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase ... 471 e-133
TC225900 homologue to UP|ODPA_PEA (P52902) Pyruvate dehydrogenas... 390 e-109
TC233409 weakly similar to UP|ODPA_SMIMA (P52900) Pyruvate dehyd... 373 e-104
TC225899 similar to UP|O48685 (O48685) Pyruvate dehydrogenase E1... 296 1e-80
TC225897 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase ... 240 7e-64
BF069526 weakly similar to SP|P52902|ODPA Pyruvate dehydrogenase... 197 5e-51
TC221704 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase ... 162 3e-40
TC205336 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto... 145 3e-35
TC206698 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase ... 115 3e-26
TC206696 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1... 92 3e-19
TC206697 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1... 79 3e-15
BE057179 similar to GP|28465343|dbj pyruvate dehydrogenase E1alp... 76 3e-14
TC206695 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1... 67 2e-11
AW133065 similar to SP|P52903|ODPA_ Pyruvate dehydrogenase E1 co... 66 2e-11
TC235074 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto... 49 4e-06
TC226476 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-ph... 42 5e-04
TC215436 similar to UP|Q9LZY8 (Q9LZY8) Transketolase-like protei... 32 0.38
TC218834 UP|Q8I727 (Q8I727) TcC31.32, partial (8%) 30 1.9
TC216950 30 2.5
TC232164 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto... 29 4.2
>TC225898 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (PDHE1-A) ,
partial (62%)
Length = 1250
Score = 471 bits (1213), Expect = e-133
Identities = 223/248 (89%), Positives = 238/248 (95%)
Frame = +1
Query: 148 GCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQ 207
GCSKGKGGSMHFYRKEGGFYGGHGIVGAQ+PLG GLAF QKY KD NVTF++YGDGAANQ
Sbjct: 1 GCSKGKGGSMHFYRKEGGFYGGHGIVGAQVPLGCGLAFAQKYCKDENVTFSMYGDGAANQ 180
Query: 208 GQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLA 267
GQLFEALNI+ALWDLP+ILVCENNHYGMGTAEWR+AKSP+YYKRGDY PGLKVDGMD LA
Sbjct: 181 GQLFEALNISALWDLPSILVCENNHYGMGTAEWRAAKSPSYYKRGDYVPGLKVDGMDALA 360
Query: 268 VKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRK 327
VKQACKFAKE ALKNGP+ILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRK
Sbjct: 361 VKQACKFAKEFALKNGPIILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRK 540
Query: 328 LVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
L+L H+I+TEKELKDIEKE RK+VDEAIAKAKESQMP+PSDLFTNVYVKG GVEACGADR
Sbjct: 541 LLLTHEIATEKELKDIEKEIRKEVDEAIAKAKESQMPEPSDLFTNVYVKGFGVEACGADR 720
Query: 388 KEVKATLP 395
KE++ATLP
Sbjct: 721 KELRATLP 744
>TC225900 homologue to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1
component alpha subunit, mitochondrial precursor
(PDHE1-A) , partial (54%)
Length = 1042
Score = 390 bits (1002), Expect = e-109
Identities = 189/206 (91%), Positives = 197/206 (94%)
Frame = +2
Query: 190 NKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYY 249
+KD +VTF +YGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWR+AKSPAYY
Sbjct: 29 SKDESVTFAMYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYY 208
Query: 250 KRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTR 309
KRGDY PGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTR
Sbjct: 209 KRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTR 388
Query: 310 DEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDL 369
DEISGVRQERDPIERVRKLVL+HDI+ EKELKDIEKE RK+VDEAIAKAKES MPDPSDL
Sbjct: 389 DEISGVRQERDPIERVRKLVLSHDIAAEKELKDIEKEVRKEVDEAIAKAKESPMPDPSDL 568
Query: 370 FTNVYVKGLGVEACGADRKEVKATLP 395
FTNVYVKG GVE GADRKEV+ATLP
Sbjct: 569 FTNVYVKGYGVETFGADRKEVRATLP 646
>TC233409 weakly similar to UP|ODPA_SMIMA (P52900) Pyruvate dehydrogenase E1
component alpha subunit, mitochondrial precursor
(PDHE1-A) (Fragment) , partial (66%)
Length = 1139
Score = 373 bits (958), Expect = e-104
Identities = 189/345 (54%), Positives = 239/345 (68%), Gaps = 2/345 (0%)
Frame = +3
Query: 34 STETLTVETSIPFTSHNCEP-PSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIR 92
ST TL + P+ H + PS V+T +L+S F M +R+MEI AD +YKAK I+
Sbjct: 84 STPTLKFQVK-PYDLHRLDKGPSLEVETNKEDLVSAFKTMYEIRQMEIIADKMYKAKKIQ 260
Query: 93 GFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKG 152
GF HLY+G+EAV G+E+ I D VITAYRDH L RGG +VF+EL G+ G S+G
Sbjct: 261 GFLHLYNGEEAVVTGIESGIRPDDHVITAYRDHGFILTRGGNAEQVFAELQGKVTGTSRG 440
Query: 153 KGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFE 212
KGGSMH Y KEG FYGG+GIVGAQ P+G GLAF KYNK V T YGDG+ANQGQLFE
Sbjct: 441 KGGSMHMYSKEGKFYGGNGIVGAQTPVGAGLAFAAKYNKTDQVCITCYGDGSANQGQLFE 620
Query: 213 ALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQAC 272
A N+AALW LP I VCENN YGMGT+ R+A S +Y RGD+ PG+++DGMD LAVK A
Sbjct: 621 AYNMAALWKLPVIFVCENNKYGMGTSIDRAAASTTFYTRGDFVPGIRIDGMDYLAVKNAT 800
Query: 273 KFAKEHA-LKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
KFA ++ GPL+LEM TYRY GHSMSDPG TYRTR+E+ VR RDPI VRK +
Sbjct: 801 KFAADYCRAGQGPLVLEMVTYRYVGHSMSDPGITYRTREEVDAVRASRDPINLVRKKITD 980
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVK 376
+++ TE EL +IE +K+++ A +A E+ P+ + +T VY++
Sbjct: 981 NNLLTEDELTEIENGIKKELNAAAQRADEAPYPEIKETYTQVYIE 1115
>TC225899 similar to UP|O48685 (O48685) Pyruvate dehydrogenase E1 alpha
subunit , partial (40%)
Length = 700
Score = 296 bits (758), Expect = 1e-80
Identities = 143/185 (77%), Positives = 162/185 (87%)
Frame = +3
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR + S LLKP ++R RS+S+SS + LT+ETS+PFTSHNC+ PS V+T
Sbjct: 147 MALSRVVAQSSQSNLLKPLASYLSLRRRSVSTSS-DPLTIETSVPFTSHNCDAPSRAVET 323
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
+++EL +FF+DM LMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI RKDCVIT
Sbjct: 324 SSAELFAFFHDMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITRKDCVIT 503
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHCTFL RGGTL+E+FSELMGR+DGCSKGKGGSMHFYRKEGGFYGGHGIVGAQ+PLG
Sbjct: 504 AYRDHCTFLARGGTLIEIFSELMGRRDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQVPLG 683
Query: 181 VGLAF 185
GLAF
Sbjct: 684 CGLAF 698
>TC225897 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (PDHE1-A) ,
partial (32%)
Length = 820
Score = 240 bits (613), Expect = 7e-64
Identities = 117/129 (90%), Positives = 124/129 (95%)
Frame = +1
Query: 267 AVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVR 326
AVKQACKFAKE ALKNGP+ILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVR
Sbjct: 1 AVKQACKFAKEFALKNGPIILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVR 180
Query: 327 KLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGAD 386
KL+L H+I+TEKELKDIEKE RK+VDEAIAKAKESQMP+PSDLFTNVYVKG GVEACGAD
Sbjct: 181 KLLLTHEIATEKELKDIEKEIRKEVDEAIAKAKESQMPEPSDLFTNVYVKGFGVEACGAD 360
Query: 387 RKEVKATLP 395
RKEV+A LP
Sbjct: 361 RKEVRAILP 387
>BF069526 weakly similar to SP|P52902|ODPA Pyruvate dehydrogenase E1
component alpha subunit mitochondrial precursor (EC
1.2.4.1) (PDHE1-A)., partial (39%)
Length = 469
Score = 197 bits (502), Expect = 5e-51
Identities = 97/156 (62%), Positives = 109/156 (69%)
Frame = +2
Query: 102 EAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYR 161
EA A M+A + RKDC+I YR HCT L GGTL+ V ELM R CS GK G MH YR
Sbjct: 2 EAFAGCMKANLTRKDCLIITYRYHCTLLTLGGTLIYVLPELMRRHHRCSNGKRGCMHLYR 181
Query: 162 KEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWD 221
E G GGH I GAQ+PL L F QKY D NV ++YGD AA GQ FEALNI ALWD
Sbjct: 182 TESGLSGGHEIYGAQVPLRCILTFAQKYCIDENVMLSMYGDRAAYLGQWFEALNIYALWD 361
Query: 222 LPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPG 257
L +I+VC+NNHYGMGTAEWR+A SP+YYKR DY PG
Sbjct: 362 LASIMVCQNNHYGMGTAEWRAANSPSYYKREDYVPG 469
>TC221704 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (46%)
Length = 616
Score = 162 bits (409), Expect = 3e-40
Identities = 85/206 (41%), Positives = 117/206 (56%), Gaps = 9/206 (4%)
Frame = +2
Query: 50 NCEPPSTT-VQTTASELMSFFNDMVLMRRME-IAADSLYKAKLIRGFCHLYDGQEAVAVG 107
N +P STT + T E + + DMVL R E + A Y+ K+ GF HLY+GQEAV+ G
Sbjct: 2 NNKPKSTTNLLITKEEGLQLYEDMVLGRSFEDMCAQMYYRGKMF-GFVHLYNGQEAVSTG 178
Query: 108 MEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFY 167
+ ++DCV++ YRDH L +G V SEL G+ GCS+G+GGSMH + KE
Sbjct: 179 FINFLKKEDCVVSTYRDHVHALSKGVPARAVMSELFGKATGCSRGQGGSMHMFSKEHNLI 358
Query: 168 GGHGIVGAQIPLGVGLAFGQKYNKD-------PNVTFTLYGDGAANQGQLFEALNIAALW 220
GG + IP+ G AF KY ++ +VT +GDG N GQ +E LN+AALW
Sbjct: 359 GGFAFIAEGIPVATGAAFSSKYRREVLKEADCDHVTLAFFGDGTCNNGQFYECLNMAALW 538
Query: 221 DLPAILVCENNHYGMGTAEWRSAKSP 246
LP + V ENN + +G + R+ P
Sbjct: 539 KLPIVFVVENNLWAIGMSHLRATSDP 616
>TC205336 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein, partial
(93%)
Length = 1642
Score = 145 bits (366), Expect = 3e-35
Identities = 95/326 (29%), Positives = 158/326 (48%), Gaps = 6/326 (1%)
Frame = +1
Query: 55 STTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLY---DGQEAVAVGMEAA 111
S VQ + + ++DMV ++ M D+++ +G Y G+EAV + AA
Sbjct: 451 SNYVQVSKEMGVKMYSDMVTLQTM----DNIFYEVQRQGRISFYLTQMGEEAVNIASAAA 618
Query: 112 INRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHG 171
+ D ++ YR+ L RG TL + + G KG+ +H+ + ++
Sbjct: 619 LAPDDIILPQYREPGVLLWRGFTLQQFVHQCFGNTHDFGKGRQMPIHYGSNQHNYFTVSS 798
Query: 172 IVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENN 231
+ Q+P VG A+ K + T GDGA ++G A+N AA+ + P + +C NN
Sbjct: 799 PIATQLPQAVGAAYSLKMDGKSACAVTFCGDGATSEGDFHAAMNFAAVMEAPVVFICRNN 978
Query: 232 HYGMGTAEWRSAKSPAYYKRGDYAP--GLKVDGMDVLAVKQACKFAKEHALK-NGPLILE 288
+ + T +S +G ++VDG D LAV A A+E A+K P+++E
Sbjct: 979 GWAISTPVEDQFRSDGIVVKGKAYGIWSIRVDGNDALAVYSAVHTAREIAIKEKRPVLIE 1158
Query: 289 MDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEAR 348
TYR HS SD + YR DEI + R+P+ R ++ V + ++K+ ++ R
Sbjct: 1159ALTYRVGHHSTSDDSTKYRGTDEIEYWKTARNPVNRFKRWVERNGWWSDKDELELRSSVR 1338
Query: 349 KQVDEAIAKAKESQMPDPSDLFTNVY 374
KQ+ AI A+++Q P DLF +VY
Sbjct: 1339KQLMHAIQVAEKAQKPPLQDLFNDVY 1416
>TC206698 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (31%)
Length = 523
Score = 115 bits (288), Expect = 3e-26
Identities = 56/134 (41%), Positives = 77/134 (56%), Gaps = 7/134 (5%)
Frame = +3
Query: 86 YKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGR 145
Y+ K+ GF HLY+GQEAV+ G + ++D V++ YRDH L +G EV SEL G+
Sbjct: 15 YRGKMF-GFVHLYNGQEAVSTGFIKLLKKEDSVVSTYRDHVHALSKGVPSREVMSELFGK 191
Query: 146 KDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKD-------PNVTFT 198
GC +G+GGSMH + KE GG +G IP+ G AF KY ++ +VT
Sbjct: 192 ATGCCRGQGGSMHMFSKEHNLLGGFAFIGEGIPVATGAAFSSKYRREVLKQADCDHVTLA 371
Query: 199 LYGDGAANQGQLFE 212
+GDG N GQ +E
Sbjct: 372 FFGDGTCNNGQFYE 413
>TC206696 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (43%)
Length = 765
Score = 92.4 bits (228), Expect = 3e-19
Identities = 53/151 (35%), Positives = 88/151 (58%), Gaps = 8/151 (5%)
Frame = +3
Query: 222 LPAILVCENNHYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKEHA 279
LP + V ENN + +G + R+ P +K+G PG+ VDGMDVL V++ K A A
Sbjct: 9 LPIVFVVENNLWAIGMSHLRATSDPQIWKKGPAFGMPGVHVDGMDVLQVREVAKEAVGRA 188
Query: 280 LK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQE-----RDPIERVRKLVLAHD 333
+ GP ++E +TYR+ GHS++DP DE+ ++ RDPI ++K + ++
Sbjct: 189 RRVEGPTLVECETYRFRGHSLADP-------DELRDPAEKEHYAVRDPITALKKYLFENN 347
Query: 334 ISTEKELKDIEKEARKQVDEAIAKAKESQMP 364
++ E+ELK IEK+ + +++A+ A S +P
Sbjct: 348 LANEQELKTIEKKIDEILEDAVEVAD*SPLP 440
>TC206697 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (33%)
Length = 566
Score = 79.3 bits (194), Expect = 3e-15
Identities = 50/134 (37%), Positives = 75/134 (55%), Gaps = 4/134 (2%)
Frame = +3
Query: 260 VDGMDVLAVKQACKFAKEHALKN-GPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQE 318
VDGMDVL V++ K A E A + GP ++E +TYR+ GHS++DP RD
Sbjct: 6 VDGMDVLKVREVAKEAIERARRGEGPTLVECETYRFRGHSLADPDEL---RDPAEKAHYA 176
Query: 319 -RDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYV-- 375
RDPI ++K ++ + +++E+E K IEK+ + V++A+ A ES L NV+
Sbjct: 177 GRDPISALKKYMIENKLASEQEFKTIEKKIEEIVEDAVEFADESPHSPRIQLLENVFADP 356
Query: 376 KGLGVEACGADRKE 389
KG G+ G R E
Sbjct: 357 KGFGIGPDGNYRCE 398
>BE057179 similar to GP|28465343|dbj pyruvate dehydrogenase E1alpha subunit
{Beta vulgaris}, partial (10%)
Length = 167
Score = 75.9 bits (185), Expect = 3e-14
Identities = 37/55 (67%), Positives = 42/55 (76%)
Frame = +1
Query: 334 ISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADRK 388
I+ EKELKDIEKE RK+ D+ + +K MPDPSDL TNVYVKG GVE GADRK
Sbjct: 1 IAAEKELKDIEKEVRKKSDQTLG*SKGKSMPDPSDLSTNVYVKGYGVETFGADRK 165
>TC206695 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (29%)
Length = 612
Score = 66.6 bits (161), Expect = 2e-11
Identities = 38/115 (33%), Positives = 67/115 (58%), Gaps = 7/115 (6%)
Frame = +1
Query: 282 NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQE-----RDPIERVRKLVLAHDIST 336
+GP ++E +TYR+ GHS++DP DE+ ++ RDPI +++ ++ ++++
Sbjct: 22 DGPTLVECETYRFRGHSLADP-------DELRDPAEKEHYAGRDPITALKQYLIENNLAN 180
Query: 337 EKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYV--KGLGVEACGADRKE 389
E+ELK IEK+ + +++A+ A S +P S L NV+ KG G+ G R E
Sbjct: 181 EQELKAIEKKIDEILEDAVEFADSSPLPPRSQLLENVFADPKGFGIGPDGRYRCE 345
>AW133065 similar to SP|P52903|ODPA_ Pyruvate dehydrogenase E1 component
alpha subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)., partial (10%)
Length = 121
Score = 66.2 bits (160), Expect = 2e-11
Identities = 31/40 (77%), Positives = 35/40 (87%)
Frame = -1
Query: 355 IAKAKESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATL 394
IA AK+ QMP+P DLFTNVYV G GVEACGADRKE++ATL
Sbjct: 121 IATAKKVQMPEPCDLFTNVYVTGSGVEACGADRKELRATL 2
>TC235074 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein, partial
(31%)
Length = 519
Score = 48.9 bits (115), Expect = 4e-06
Identities = 22/60 (36%), Positives = 32/60 (52%)
Frame = +2
Query: 172 IVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENN 231
+ QI VG A+ K +K T +GDG +++G ALN AA+ + P I +C NN
Sbjct: 329 VYSTQISHAVGAAYSLKMDKKDACAVTYFGDGGSSEGDFHAALNFAAVLEAPVIFICRNN 508
Score = 29.3 bits (64), Expect = 3.2
Identities = 18/62 (29%), Positives = 27/62 (43%)
Frame = +3
Query: 106 VGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGG 165
V AA+ D V YR+ L RG TL E ++L KG+ H+ K+
Sbjct: 3 VASAAALAMDDVVFPQYREAGVLLWRGFTLQEFANQLFSNIYDYGKGRQMPAHYGSKKHN 182
Query: 166 FY 167
++
Sbjct: 183 YF 188
>TC226476 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-phosphate
synthase 1 precursor, partial (85%)
Length = 2000
Score = 42.0 bits (97), Expect = 5e-04
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 12/92 (13%)
Frame = +3
Query: 152 GKGGSMHFYRKEGGFYG------------GHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
G+ MH R+ G G G G I G+G+A G+ N +
Sbjct: 429 GRRDQMHTMRQTNGLSGFTKRSESEFDCFGTGHSSTTISAGLGMAVGRDLKGRKNNVVAV 608
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENN 231
GDGA GQ +EA+N A D I++ +N
Sbjct: 609 IGDGAMTAGQAYEAMNNAGYLDSDMIVILNDN 704
>TC215436 similar to UP|Q9LZY8 (Q9LZY8) Transketolase-like protein, partial
(65%)
Length = 1625
Score = 32.3 bits (72), Expect = 0.38
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 11/73 (15%)
Frame = +2
Query: 171 GIVGAQIPLGVGLAFGQK-----YNKDPN-----VTFTLYGDGAANQGQLFEALNIAALW 220
G +G I VGLA +K +NK N T+ + GDG +G EA ++A W
Sbjct: 614 GPLGQGIANAVGLALAEKHLAARFNKPDNEIVDHYTYVILGDGCQMEGISNEACSLAGHW 793
Query: 221 DL-PAILVCENNH 232
L I + ++NH
Sbjct: 794 GLGKLIALYDDNH 832
>TC218834 UP|Q8I727 (Q8I727) TcC31.32, partial (8%)
Length = 1405
Score = 30.0 bits (66), Expect = 1.9
Identities = 24/95 (25%), Positives = 44/95 (46%)
Frame = +3
Query: 273 KFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAH 332
K +E L+ L+ E+D R T E+ RQ IER + L H
Sbjct: 375 KHLEEATLREADLLQELDRER-----------TAEMEKELE--RQRLLEIERAKTKELRH 515
Query: 333 DISTEKELKDIEKEARKQVDEAIAKAKESQMPDPS 367
++ EKE + ++E ++++++A + + S+ PS
Sbjct: 516 NLDMEKE-RQTQRELQREIEQAESGLRPSRRDFPS 617
>TC216950
Length = 873
Score = 29.6 bits (65), Expect = 2.5
Identities = 16/37 (43%), Positives = 21/37 (56%)
Frame = +2
Query: 327 KLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQM 363
+ V H E ELK IE+E K+V+EAI K E +
Sbjct: 182 QFVCFHRRQLEAELKLIEEETAKRVEEAIRKRVEESL 292
>TC232164 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein, partial
(19%)
Length = 764
Score = 28.9 bits (63), Expect = 4.2
Identities = 20/79 (25%), Positives = 36/79 (45%)
Frame = +1
Query: 122 YRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGV 181
YR+ L RG TL E ++L K KG+ H+ K+ ++ V + I L +
Sbjct: 64 YREAGVLLWRGFTLQEFANQLFSNKYDNGKGRQIPAHYGSKKHNYF----TVASTIALSI 231
Query: 182 GLAFGQKYNKDPNVTFTLY 200
+ + Q ++ + F L+
Sbjct: 232 EIFYIQGFDVSLYILFYLF 288
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,736,064
Number of Sequences: 63676
Number of extensions: 231804
Number of successful extensions: 1203
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1181
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1188
length of query: 395
length of database: 12,639,632
effective HSP length: 99
effective length of query: 296
effective length of database: 6,335,708
effective search space: 1875369568
effective search space used: 1875369568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146588.4


