

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.12 - phase: 0 /pseudo
(840 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
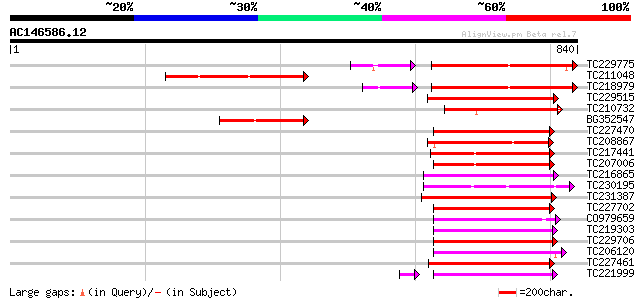
Score E
Sequences producing significant alignments: (bits) Value
TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 214 9e-68
TC211048 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associate... 238 6e-63
TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associate... 213 1e-61
TC229515 similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (20%) 172 4e-43
TC210732 similar to UP|Q39143 (Q39143) Light repressible recepto... 167 2e-41
BG352547 similar to GP|9837280|gb| leaf senescence-associated re... 155 7e-38
TC227470 similar to UP|Q8L7V7 (Q8L7V7) AT4g02010/T10M13_2, parti... 148 9e-36
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 147 2e-35
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 145 9e-35
TC207006 similar to UP|Q75UP2 (Q75UP2) Leucine-rich repeat recep... 144 1e-34
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 144 2e-34
TC230195 similar to UP|Q9FIU5 (Q9FIU5) Serine/threonine-specific... 143 3e-34
TC231387 similar to UP|Q9FUK3 (Q9FUK3) Cytokinin-regulated kinas... 142 5e-34
TC227702 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 141 1e-33
CO979659 140 2e-33
TC219303 homologue to UP|Q6W0C7 (Q6W0C7) Pto-like serine/threoni... 140 2e-33
TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 140 2e-33
TC206120 similar to UP|Q941F6 (Q941F6) Leucine-rich repeat recep... 140 3e-33
TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein ... 140 3e-33
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 136 5e-33
>TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (8%)
Length = 1250
Score = 214 bits (544), Expect(2) = 9e-68
Identities = 103/218 (47%), Positives = 153/218 (69%), Gaps = 3/218 (1%)
Frame = +1
Query: 626 SNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAF 685
+ L W +RL IA+DAA GL+YLHNGCKPP +HRD+K +NILL+EN AK+ADFGLS++F
Sbjct: 352 AKFLTWEDRLQIALDAAQGLEYLHNGCKPPIIHRDVKCANILLNENFQAKLADFGLSKSF 531
Query: 686 DNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESI 745
D S++ST AGT GY+DP++ + +K+D+YSFG+VLLE++TG+ A+ + + +
Sbjct: 532 PTDGGSYMSTVVAGTPGYLDPEYSISSRLTEKSDVYSFGVVLLEMVTGQPAIAK-TPDKT 708
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
HI QWV ++ GDI++I D+R + FD +S W++VEI M+S S +RP MS I+ EL
Sbjct: 709 HISQWVKSMLSNGDIKNIADSRFKEDFDTSSVWRIVEIGMASVSISPFKRPSMSYIVNEL 888
Query: 806 KECLSLDMVHRNNGRERA---IVELTSLNIASDTIPLA 840
KECL+ ++ + +GR+ +EL +LN ++ P A
Sbjct: 889 KECLTTELARKYSGRDTENSDSIELVTLNFTTELGPPA 1002
Score = 62.8 bits (151), Expect(2) = 9e-68
Identities = 43/100 (43%), Positives = 56/100 (56%), Gaps = 4/100 (4%)
Frame = +3
Query: 505 IKTSKIQLY*NCQYY**LQNYYWRRRIWKSLLW----HSTRSN*SRC*DAFTIINARL*G 560
I+ + I + * C + L W R IWKSL W HS S C DAF++ + R+*
Sbjct: 33 IQATAIYIQ*AC*DHQQLYQNSW*RWIWKSLSWIY**HS-----SSCKDAFSVSSTRI*T 197
Query: 561 I*SRGTTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQWK 600
I SRG TS+ SS*K CW+L *R +L*I+G+WK
Sbjct: 198 IPSRG*TSDESSS*KSDFSCWVLQ*RKQHRAHL*IYGKWK 317
>TC211048 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associated
receptor-like protein kinase, partial (24%)
Length = 675
Score = 238 bits (608), Expect = 6e-63
Identities = 120/211 (56%), Positives = 156/211 (73%)
Frame = +3
Query: 232 YIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEPLRRAETYQISLSKT 291
Y+HF E++ LA N+TREFNIT+N KLW+ +P Y + + I++T + + S T
Sbjct: 3 YMHFTEIQVLAKNQTREFNITLNGKLWYENESPRYHSVNTIYTTSGIS-GKLINFSFVMT 179
Query: 292 KNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAYGVTRNWQGDPCAPVNYMWEG 351
+ STLPPI+NA EIY K+F Q +T Q DVD IT IK+ YGVTR+WQGDPC+P +Y+WEG
Sbjct: 180 ETSTLPPIINAIEIYRVKEFPQQDTYQGDVDAITTIKSVYGVTRDWQGDPCSPKDYLWEG 359
Query: 352 LNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLR 411
LNC T + PRI +LNLSSSGL+G+I SI LTML+ LDLSNNSLNG +PDFL QL+
Sbjct: 360 LNC-TYPVIDSPRIITLNLSSSGLSGKIDPSILNLTMLEKLDLSNNSLNGEVPDFLSQLQ 536
Query: 412 SLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
L++LN+ NNL+G +PS L+E+SK GSLSL
Sbjct: 537 YLKILNLENNNLSGSIPSTLVEKSKEGSLSL 629
>TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associated
receptor-like protein kinase, partial (34%)
Length = 1343
Score = 213 bits (543), Expect(2) = 1e-61
Identities = 108/217 (49%), Positives = 154/217 (70%), Gaps = 2/217 (0%)
Frame = +3
Query: 626 SNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAF 685
S L+W +RL IAVDAA GL+YL NGCKPP +HRD+K +NILL+E+ AK++DFGLS+A
Sbjct: 462 STFLSWEDRLRIAVDAALGLEYLQNGCKPPIIHRDVKSTNILLNEHFQAKLSDFGLSKAI 641
Query: 686 DNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESI 745
D +SH+ST AGT GY+DP + +K+D+YSFG+VLLE+IT + + R + E
Sbjct: 642 PTDGESHVSTVLAGTPGYLDPHCHISSRLTQKSDVYSFGVVLLEIITNQPVMER-NQEKG 818
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
HI + V ++E+GDIR+I+D+RL+G +DINSAWK +EIAM+ S ERP MS+I EL
Sbjct: 819 HIRERVRSLIEKGDIRAIVDSRLEGDYDINSAWKALEIAMACVSQNPNERPIMSEIAIEL 998
Query: 806 KECLSLDMVHRN--NGRERAIVELTSLNIASDTIPLA 840
KE L++++ + R +VE S+N+ ++ +PLA
Sbjct: 999 KETLAIEIARAKHCDANPRYLVEAVSVNVDTEFMPLA 1109
Score = 42.7 bits (99), Expect(2) = 1e-61
Identities = 33/83 (39%), Positives = 46/83 (54%), Gaps = 1/83 (1%)
Frame = +2
Query: 523 QNYYWRRRIWKSLLW-HSTRSN*SRC*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCW 581
Q YW RRIW SL + +S+ S AF+I+ L I SRG TS+ SS K +P W
Sbjct: 197 QYNYW*RRIWNSLPGLYR*QSSCSE--SAFSIVCKWLSTISSRG*TSSQSSSQKLNIPYW 370
Query: 582 IL**R*NQSINL*IHGQWKSPTT 604
+L *R ++*++G W+ T
Sbjct: 371 LLQ*RNQ*GSHI*VYG*WELTRT 439
>TC229515 similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (20%)
Length = 919
Score = 172 bits (437), Expect = 4e-43
Identities = 85/195 (43%), Positives = 122/195 (61%), Gaps = 1/195 (0%)
Frame = +1
Query: 619 YLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIAD 678
++H L+W RL IA DAA GL+YLH GC P +HRD+K NILLD NM AK++D
Sbjct: 25 HIHESSKKKNLDWLTRLRIAEDAAKGLEYLHTGCNPSIIHRDIKTGNILLDINMRAKVSD 204
Query: 679 FGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALV 738
FGLSR + D+ +HIS+ GT GY+DP++ + +K+D+YSFG+VLLELI+GKK +
Sbjct: 205 FGLSRLAEEDL-THISSIARGTVGYLDPEYYASQQLTEKSDVYSFGVVLLELISGKKPVS 381
Query: 739 RAS-GESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPD 797
G+ ++I+ W + +GD SIID L G S W+VVEIAM + RP
Sbjct: 382 SEDYGDEMNIVHWARSLTRKGDAMSIIDPSLAGNAKTESIWRVVEIAMQCVAQHGASRPR 561
Query: 798 MSQILAELKECLSLD 812
M +I+ +++ ++
Sbjct: 562 MQEIILAIQDATKIE 606
>TC210732 similar to UP|Q39143 (Q39143) Light repressible receptor protein
kinase , partial (9%)
Length = 822
Score = 167 bits (422), Expect = 2e-41
Identities = 86/186 (46%), Positives = 120/186 (64%), Gaps = 11/186 (5%)
Frame = +1
Query: 644 GLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDNDID-----------SH 692
GLDYLH+GCKPP +HRD+K +NILL E+ AKIADFGLSR F D ++
Sbjct: 208 GLDYLHHGCKPPIIHRDVKSANILLSEDFEAKIADFGLSREFRTDNQDQQFQVIHKDATY 387
Query: 693 ISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVT 752
+ GT GY+DP++ + G N+K+DIYSFGIVLLEL+TG+ A+++ + +HIL+W+
Sbjct: 388 EKSAVMGTTGYLDPEYYKLGRLNEKSDIYSFGIVLLELLTGRPAILKGN-RVMHILEWIR 564
Query: 753 PIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLD 812
P +ERGD+ IID RLQGKFD + WK + IA + ++AE+K+CL +
Sbjct: 565 PELERGDLSKIIDPRLQGKFDASXGWKALGIASHALHQPPFRDIQ*VIVIAEMKQCLXXE 744
Query: 813 MVHRNN 818
H +N
Sbjct: 745 S-HSDN 759
>BG352547 similar to GP|9837280|gb| leaf senescence-associated receptor-like
protein kinase {Phaseolus vulgaris}, partial (15%)
Length = 472
Score = 155 bits (392), Expect = 7e-38
Identities = 78/132 (59%), Positives = 101/132 (76%)
Frame = +2
Query: 311 FSQLETQQDDVDNITNIKNAYGVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNL 370
F Q +T Q DVD I IK+ YG+TR+WQGDPC+PV Y+W GLN + + NP RIT+LNL
Sbjct: 2 FQQSDTHQGDVDAIATIKSVYGMTRDWQGDPCSPVAYLWNGLNWTYRGNENP-RITTLNL 178
Query: 371 SSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSE 430
SSS L+G I S+S LTML+ LDLSNN+LNG PDFL +L+ L++++V NNLTG +PSE
Sbjct: 179 SSSELSGMIDPSMSYLTMLEKLDLSNNNLNGEGPDFLSRLQHLKIIDVDHNNLTGSIPSE 358
Query: 431 LLERSKTGSLSL 442
L+++SK G LSL
Sbjct: 359 LVKKSKEGFLSL 394
>TC227470 similar to UP|Q8L7V7 (Q8L7V7) AT4g02010/T10M13_2, partial (34%)
Length = 1061
Score = 148 bits (374), Expect = 9e-36
Identities = 76/181 (41%), Positives = 116/181 (63%), Gaps = 3/181 (1%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W+ R+ IA+DAA GL YLH +P +HRD K SNILL+ N HAK+ADFGL++
Sbjct: 19 LDWDTRMKIALDAARGLAYLHEDSQPCVIHRDFKASNILLENNFHAKVADFGLAKQAPEG 198
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA--LVRASGESIH 746
+++STR GTFGYV P++ TG+ K+D+YS+G+VLLEL+TG+K + + SG+ +
Sbjct: 199 RANYLSTRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPSGQE-N 375
Query: 747 ILQWVTPIVERGD-IRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
++ W PI+ D + + D RL G++ +V IA + +P +RP M +++ L
Sbjct: 376 LVTWARPILRDKDRLEELADPRLGGRYPKEDFVRVCTIAAACVAPEASQRPTMGEVVQSL 555
Query: 806 K 806
K
Sbjct: 556 K 558
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 147 bits (371), Expect = 2e-35
Identities = 78/194 (40%), Positives = 119/194 (61%), Gaps = 7/194 (3%)
Frame = +1
Query: 619 YLHAVENSNM----LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHA 674
+L+ V NS + L+W RL IA++AA GL+YLH PP +HRD K SNILLD+ HA
Sbjct: 250 HLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLHEHVSPPVIHRDFKSSNILLDKKFHA 429
Query: 675 KIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGK 734
K++DFGL++ + H+STR GT GYV P++ TG+ K+D+YS+G+VLLEL+TG+
Sbjct: 430 KVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALTGHLTTKSDVYSYGVVLLELLTGR 609
Query: 735 KA--LVRASGESIHILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPI 791
+ R GE + ++ W P++ +R + I+D L+G++ + +V IA P
Sbjct: 610 VPVDMKRPPGEGV-LVSWALPLLTDREKVVKIMDPSLEGQYSMKEVVQVAAIAAMCVQPE 786
Query: 792 EVERPDMSQILAEL 805
RP M+ ++ L
Sbjct: 787 ADYRPLMADVVQSL 828
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 145 bits (365), Expect = 9e-35
Identities = 70/186 (37%), Positives = 121/186 (64%), Gaps = 3/186 (1%)
Frame = +2
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
E+ L W ER IA+ +A GL YLH+ C P +HRD+K +NILLDE A + DFGL++
Sbjct: 428 ESQPPLGWPERKRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAK 607
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA--LVR-A 740
D D+H++T GT G++ P++ TG +++K D++ +G++LLELITG++A L R A
Sbjct: 608 LMDYK-DTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLA 784
Query: 741 SGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQ 800
+ + + +L WV +++ + +++DA LQG ++ +++++A+ T +ERP MS+
Sbjct: 785 NDDDVMLLDWVKGLLKDRKLETLVDADLQGSYNDEEVEQLIQVALLCTQGSPMERPKMSE 964
Query: 801 ILAELK 806
++ L+
Sbjct: 965 VVRMLE 982
>TC207006 similar to UP|Q75UP2 (Q75UP2) Leucine-rich repeat receptor-like
kinase, partial (41%)
Length = 1080
Score = 144 bits (364), Expect = 1e-34
Identities = 74/180 (41%), Positives = 114/180 (63%), Gaps = 2/180 (1%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W R IA+ A GL YLH C P +HRD+K +NILLDE+ A + DFGL++ D+
Sbjct: 99 LDWTRRKRIALGTARGLVYLHEQCDPKIIHRDVKAANILLDEDFEAVVGDFGLAKLLDHR 278
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
DSH++T GT G++ P++ TG +++K D++ FGI+LLELITG KAL RA+ +
Sbjct: 279 -DSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGHKALDFGRAANQKGV 455
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+L WV + + G + ++D L+G FD+ ++V++A+ T RP MS++L L+
Sbjct: 456 MLDWVKKLHQDGRLSQMVDKDLKGNFDLIELEEMVQVALLCTQFNPSHRPKMSEVLKMLE 635
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 144 bits (363), Expect = 2e-34
Identities = 77/201 (38%), Positives = 121/201 (59%), Gaps = 1/201 (0%)
Frame = +1
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
N A A E+ L+W R I V A GL YLH + +HRD+K +N+LLD+++
Sbjct: 232 NSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIVHRDIKATNVLLDQDL 411
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
+ KI+DFGL++ D + ++HISTR AGTFGY+ P++ G K D+YSFGIV LE+I
Sbjct: 412 NPKISDFGLAK-LDEEDNTHISTRIAGTFGYMAPEYAMHGYLTDKADVYSFGIVALEIIN 588
Query: 733 GKKALV-RASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPI 791
G+ + R ES +L+W + E+GDI ++D RL +F+ A ++++A+ T+
Sbjct: 589 GRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKEEALVMIKVALLCTNVT 768
Query: 792 EVERPDMSQILAELKECLSLD 812
RP MS +++ L+ + +D
Sbjct: 769 AALRPTMSSVVSMLEGKIVVD 831
>TC230195 similar to UP|Q9FIU5 (Q9FIU5) Serine/threonine-specific protein
kinase-like protein, partial (32%)
Length = 889
Score = 143 bits (361), Expect = 3e-34
Identities = 77/225 (34%), Positives = 127/225 (56%)
Frame = +3
Query: 613 NFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENM 672
+ +++ Y +N L+W+ RL+IA+D A GL+YLH+G PP +HRD+K NILLD++M
Sbjct: 3 SLDSHLYADLGKNHKPLSWDLRLSIALDVARGLEYLHHGASPPVVHRDIKSCNILLDQSM 182
Query: 673 HAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELIT 732
AK+ DFGLSR ++ ++ GTFGYVDP++ T KK+D+YSFG++L ELIT
Sbjct: 183 RAKVTDFGLSRP---EMIKPRTSNVRGTFGYVDPEYLSTRTFTKKSDVYSFGVLLFELIT 353
Query: 733 GKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIE 792
G+ G ++ V + I+D +L GK+D+++ + +A + +
Sbjct: 354 GRNP---QQGLMEYVKLAVMESEGKVGWEEIVDPQLNGKYDVHNLHDMASLAFKCVNEVS 524
Query: 793 VERPDMSQILAELKECLSLDMVHRNNGRERAIVELTSLNIASDTI 837
RP M +I+ EL + + + + G A ++ S+ + I
Sbjct: 525 KSRPSMCEIVQELSQ-ICKRQIKDHGGTSPAALKEVSIEVGQTEI 656
>TC231387 similar to UP|Q9FUK3 (Q9FUK3) Cytokinin-regulated kinase 1, partial
(29%)
Length = 765
Score = 142 bits (359), Expect = 5e-34
Identities = 75/206 (36%), Positives = 127/206 (61%), Gaps = 5/206 (2%)
Frame = +3
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N + +LH +++S +++W R+ +A+DAA G++YLH PP +HRD+K +NILLD
Sbjct: 51 YMDNGSLSDHLHKLQSSALMSWAVRIKVALDAARGIEYLHQYATPPIIHRDIKSANILLD 230
Query: 670 ENMHAKIADFGLSRAFDN--DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVL 727
AK++DFGLS + D D+H+S AGT GY+DP++ R + K+D+YSFG+VL
Sbjct: 231 AKWTAKVSDFGLSLMGPDPEDEDAHLSLLAAGTVGYMDPEYYRLQHLTPKSDVYSFGVVL 410
Query: 728 LELITGKKALVR-ASGESIHILQWVTPIVERGDIRSIIDARL--QGKFDINSAWKVVEIA 784
LEL++G KA+ + +G +++ +V P + + +I ++D R+ F+I + V +A
Sbjct: 411 LELLSGYKAIHKNENGVPRNVVDFVVPFIFQDEIHRVLDRRVAPPTPFEIEAVAYVGYLA 590
Query: 785 MSSTSPIEVERPDMSQILAELKECLS 810
+RP MSQ++ L+ L+
Sbjct: 591 ADCVRLEGRDRPTMSQVVNNLERALA 668
>TC227702 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (21%)
Length = 1142
Score = 141 bits (356), Expect = 1e-33
Identities = 69/179 (38%), Positives = 114/179 (63%), Gaps = 1/179 (0%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W R+ I + A GL YLH + +HRD+K +N+LLD+++HAKI+DFGL++ D +
Sbjct: 55 LDWPRRMQICLGIAKGLAYLHEESRLKIVHRDIKATNVLLDKHLHAKISDFGLAK-LDEE 231
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGK-KALVRASGESIHI 747
++HISTR AGT GY+ P++ G K D+YSFGIV LE+++GK R E +++
Sbjct: 232 ENTHISTRIAGTIGYMAPEYAMRGYLTDKADVYSFGIVALEIVSGKSNTNYRPKEEFVYL 411
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
L W + E+G++ ++D L K+ A +++++A+ T+P RP MS +++ L+
Sbjct: 412 LDWAYVLQEQGNLLELVDPSLGSKYSSEEAMRMLQLALLCTNPSPTLRPCMSSVVSMLE 588
>CO979659
Length = 731
Score = 140 bits (354), Expect = 2e-33
Identities = 72/190 (37%), Positives = 113/190 (58%), Gaps = 2/190 (1%)
Frame = -1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
++W R+ IA AA GL+YLHNG P ++RD K SNILLDEN + K++DFGL++ +
Sbjct: 647 MDWKNRMKIAEGAARGLEYLHNGADPAIIYRDFKSSNILLDENFNPKLSDFGLAKIGPKE 468
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASG-ESIHI 747
+ H++TR GTFGY P++ +G + K+DIYSFG+VLLE+ITG++ A G E ++
Sbjct: 467 GEEHVATRVMGTFGYCAPEYAASGQLSTKSDIYSFGVVLLEIITGRRVFDTARGTEEQNL 288
Query: 748 LQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+ W P+ +R + D L+G+F + ++ + +A E PD + ++
Sbjct: 287 IDWAQPLFKDRTKFTLMADPLLKGQFPVKGLFQALAVAAMCLQ----EEPDTRPYMDDVV 120
Query: 807 ECLSLDMVHR 816
L+ VHR
Sbjct: 119 TALAHLAVHR 90
>TC219303 homologue to UP|Q6W0C7 (Q6W0C7) Pto-like serine/threonine kinase
(Fragment), partial (73%)
Length = 1006
Score = 140 bits (353), Expect = 2e-33
Identities = 72/184 (39%), Positives = 107/184 (58%), Gaps = 1/184 (0%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W ERL I + +A GL YLH G +HRD+K +NILLDEN+ AK+ADFGLS+
Sbjct: 81 LSWKERLEICIGSARGLHYLHTGYAKAVIHRDVKSANILLDENLMAKVADFGLSKTGPEI 260
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIHI 747
+H+ST G+FGY+DP++ R +K+D+YSFG+VL E++ + + E +++
Sbjct: 261 DQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCARPVIDPTLPREMVNL 440
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
+W ++G + IID L GK +S K E A + V+RP M +L L+
Sbjct: 441 AEWALKWQKKGQLEQIIDQTLAGKIRPDSLRKFGETAEKCLADYGVDRPSMGDVLWNLEY 620
Query: 808 CLSL 811
L L
Sbjct: 621 ALQL 632
>TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (50%)
Length = 1162
Score = 140 bits (353), Expect = 2e-33
Identities = 75/186 (40%), Positives = 116/186 (62%), Gaps = 3/186 (1%)
Frame = +2
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L W+ R+ IA+ AA GL +LH + P ++RD K SNILLD +AK++DFGL++
Sbjct: 83 LPWSIRMKIALGAAKGLAFLHEEAQRPIIYRDFKTSNILLDAEYNAKLSDFGLAKDGPEG 262
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
+H+STR GT+GY P++ TG+ K+D+YSFG+VLLE++TG++++ R +GE +
Sbjct: 263 EKTHVSTRVMGTYGYAAPEYVMTGHLTSKSDVYSFGVVLLEMLTGRRSIDKKRPNGEH-N 439
Query: 747 ILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
+++W P++ +R IID RL+G F + A K +A S RP MS+++ L
Sbjct: 440 LVEWARPVLGDRRMFYRIIDPRLEGHFSVKGAQKAALLAAQCLSRDPKSRPLMSEVVRAL 619
Query: 806 KECLSL 811
K SL
Sbjct: 620 KPLPSL 637
>TC206120 similar to UP|Q941F6 (Q941F6) Leucine-rich repeat receptor-like
kinase F21M12.36, partial (18%)
Length = 1037
Score = 140 bits (352), Expect = 3e-33
Identities = 78/201 (38%), Positives = 120/201 (58%), Gaps = 5/201 (2%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W R IAV AA GL+YLH+GC+ P +HRD+K SNILLDE + +IADFGL++ +
Sbjct: 138 LDWETRYEIAVGAAKGLEYLHHGCEKPVIHRDVKSSNILLDEFLKPRIADFGLAKVIQAN 317
Query: 689 IDSHISTRP-AGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHI 747
+ ST AGT GY+ P++ T N+K+D+YSFG+VL+EL+TGK+ GE+ I
Sbjct: 318 VVKDSSTHVIAGTHGYIAPEYGYTYKVNEKSDVYSFGVVLMELVTGKRPTEPEFGENKDI 497
Query: 748 LQWV-TPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+ WV + +RS +D+R+ + A KV+ A+ T + RP M ++ +L+
Sbjct: 498 VSWVHNKARSKEGLRSAVDSRIPEMY-TEEACKVLRTAVLCTGTLPALRPTMRAVVQKLE 674
Query: 807 E---CLSLDMVHRNNGRERAI 824
+ C + +V + E+ I
Sbjct: 675 DAEPCKLVGIVITKDDSEKKI 737
>TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (59%)
Length = 1204
Score = 140 bits (352), Expect = 3e-33
Identities = 75/189 (39%), Positives = 115/189 (60%), Gaps = 3/189 (1%)
Frame = +2
Query: 621 HAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFG 680
H S L W+ R+ IA+ AA GL +LH + P ++RD K SNILLD +AK++DFG
Sbjct: 128 HLFRRSLPLPWSIRMKIALGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFG 307
Query: 681 LSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--V 738
L++ +H+STR GT+GY P++ TG+ ++D+YSFG+VLLE++TG++++
Sbjct: 308 LAKDGPEGDKTHVSTRVMGTYGYAAPEYVMTGHLTSRSDVYSFGVVLLEMLTGRRSMDKN 487
Query: 739 RASGESIHILQWVTP-IVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPD 797
R +GE ++++W P + ER +ID RL+G F I A K +A S RP
Sbjct: 488 RPNGEH-NLVEWARPHLGERRRFYKLIDPRLEGHFSIKGAQKAAHLAAHCLSRDPKARPL 664
Query: 798 MSQILAELK 806
MS+++ LK
Sbjct: 665 MSEVVEALK 691
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 136 bits (342), Expect(2) = 5e-33
Identities = 72/184 (39%), Positives = 108/184 (58%), Gaps = 1/184 (0%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W +RL I + AA GL YLH G +HRD+K +NIL+D+N AK+ADFGLS+
Sbjct: 247 LSWKQRLEICIGAARGLHYLHTGASQSIIHRDVKTTNILVDDNFVAKVADFGLSKTGPAL 426
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRA-SGESIHI 747
+H+ST G+FGY+DP++ R +K+D+YSFG+VL+E++ + AL E ++I
Sbjct: 427 DQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLMEVLCTRPALNPVLPREQVNI 606
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
+W ++G + I+D L GK + S K E A + V+RP M +L L+
Sbjct: 607 AEWAMSWQKKGMLDQIMDQNLVGKVNPASLKKFGETAEKCLAEYGVDRPSMGDVLWNLEY 786
Query: 808 CLSL 811
L L
Sbjct: 787 ALQL 798
Score = 24.3 bits (51), Expect(2) = 5e-33
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +3
Query: 578 LPCWIL**R*NQSINL*IHGQWKSPTTFI 606
+P W+L * L*IHG W S +F+
Sbjct: 141 VPHWLLR*EIRNDSCL*IHG*WTSQESFV 227
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.343 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,635,721
Number of Sequences: 63676
Number of extensions: 512224
Number of successful extensions: 6680
Number of sequences better than 10.0: 994
Number of HSP's better than 10.0 without gapping: 4747
Number of HSP's successfully gapped in prelim test: 150
Number of HSP's that attempted gapping in prelim test: 1162
Number of HSP's gapped (non-prelim): 5314
length of query: 840
length of database: 12,639,632
effective HSP length: 105
effective length of query: 735
effective length of database: 5,953,652
effective search space: 4375934220
effective search space used: 4375934220
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146586.12


