

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.9 - phase: 0
(724 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
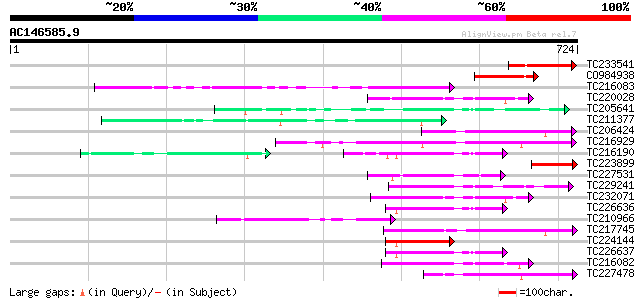
Score E
Sequences producing significant alignments: (bits) Value
TC233541 weakly similar to UP|Q96337 (Q96337) AMP-binding protei... 144 1e-34
CO984938 117 1e-26
TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 107 1e-23
TC220028 weakly similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA li... 99 5e-21
TC205641 UP|Q8S5C1 (Q8S5C1) 4-coumarate:CoA ligase isoenzyme 2 ... 99 9e-21
TC211377 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synth... 98 1e-20
TC206424 similar to UP|Q8LKS6 (Q8LKS6) Long chain acyl-CoA synth... 93 4e-19
TC216929 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partia... 92 8e-19
TC216190 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ... 92 8e-19
TC223899 similar to UP|Q8LRT1 (Q8LRT1) Acyl-CoA synthetase-like ... 90 4e-18
TC227531 similar to UP|Q84P23 (Q84P23) 4-coumarate-CoA ligase-li... 89 9e-18
TC229241 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-li... 87 3e-17
TC232071 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 86 8e-17
TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL ... 83 4e-16
TC210966 similar to UP|Q96338 (Q96338) AMP-binding protein , par... 82 6e-16
TC217745 similar to UP|Q96538 (Q96538) Acyl-CoA synthetase , pa... 82 1e-15
TC224144 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ... 80 2e-15
TC226637 UP|Q8S5C2 (Q8S5C2) 4-coumarate:CoA ligase isoenzyme 3 ... 80 2e-15
TC216082 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 80 3e-15
TC227478 similar to UP|Q96537 (Q96537) Acyl CoA synthetase , pa... 80 4e-15
>TC233541 weakly similar to UP|Q96337 (Q96337) AMP-binding protein, partial
(9%)
Length = 325
Score = 144 bits (364), Expect = 1e-34
Identities = 75/87 (86%), Positives = 82/87 (94%)
Frame = +1
Query: 637 NSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNEPFTID 696
+ EEVLKVAR+LSIIDS SS+V SEEKV +LIYKELKTW SESPFQIGPILLVNEPFTID
Sbjct: 13 SKEEVLKVARKLSIIDSNSSDV-SEEKVTSLIYKELKTWTSESPFQIGPILLVNEPFTID 189
Query: 697 NGLMTPTMKIRRDRVVAKYKEQIDDLY 723
NGLMTPTMKIRRDRVVA+Y+EQID+LY
Sbjct: 190 NGLMTPTMKIRRDRVVAQYREQIDNLY 270
>CO984938
Length = 791
Score = 117 bits (294), Expect = 1e-26
Identities = 62/82 (75%), Positives = 70/82 (84%)
Frame = -1
Query: 594 IVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDS 653
+ + GENVEP ELEEAAMRSS+I QIVVIGQDKRRLGA+IVPN EEVLK ARE SIIDS
Sbjct: 770 VSIREGENVEPGELEEAAMRSSLIHQIVVIGQDKRRLGAVIVPNKEEVLKAARESSIIDS 591
Query: 654 ISSNVVSEEKVLNLIYKELKTW 675
SS+ S+EKV +LIYKEL+TW
Sbjct: 590 NSSD-ASQEKVTSLIYKELRTW 528
>TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (80%)
Length = 1411
Score = 107 bits (268), Expect = 1e-23
Identities = 121/467 (25%), Positives = 199/467 (41%), Gaps = 7/467 (1%)
Frame = +2
Query: 109 ITYNQLEQAILDYAEGLRV-IGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRS 167
+TY QL +++ A L V +G+R + + + NS + V +M+ GAI +
Sbjct: 161 LTYTQLWRSVEGVAASLSVDMGIRKGNVVLILSPNSIHFPVVCLAVMSLGAIITTTNPLN 340
Query: 168 SVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKE 227
+ E+ + S+ + LA ++ +I S I+L+ + ++ N N
Sbjct: 341 TTREIAKQIADSKPL-LAFTISDLLPKITAAAPSLP----IVLMDNDGANNN-----NNN 490
Query: 228 VPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTH 287
I + ++ ++ +QR V E + DD ATL+Y+SGTTG KGV+ +H
Sbjct: 491 NNIVATLD----------EMAKKEPVAQR-VKERVEQDDTATLLYSSGTTGPSKGVVSSH 637
Query: 288 RNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKDDLG 347
RNL+ ++ + E + F+ +P +H Y F G+ +T+ L
Sbjct: 638 RNLIAMVQIVLGRFHMEENETFICTVPMFHIYGLVA----FATGLLASGSTI----VVLS 793
Query: 348 RYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTR 407
+++ H M+ S I++ +T P+
Sbjct: 794 KFEMHDML----------SSIERFRATYLPL----------------------------- 856
Query: 408 NVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVY--SKIHSAIGLSKAGISGGGSLPL 465
P I+ +ML+ + AIK Y + +HS + SGG L
Sbjct: 857 ---VPPILVAMLN--------------NAAAIKGKYDITSLHSVL-------SGGAPLSK 964
Query: 466 EVDKFFEA--IGVKVQNGYGLTETSPVIAA--RRPRCNVIGSVGHPVQHTEFKVVDSETG 521
EV + F A V + GYGLTE++ V A+ G+ G T+ +VD E+G
Sbjct: 965 EVIEGFVAKYPNVTILQGYGLTESTGVGASTDSLEESRRYGTAGLLSPATQAMIVDPESG 1144
Query: 522 EVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWI 568
+ LP G L +RGP +M GY+ N AT LD GWL TGD+G+I
Sbjct: 1145QSLPVNRTGELWLRGPTIMKGYFSNEEATTSTLDSKGWLRTGDIGYI 1285
>TC220028 weakly similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like
protein, partial (30%)
Length = 988
Score = 99.4 bits (246), Expect = 5e-21
Identities = 76/219 (34%), Positives = 106/219 (47%), Gaps = 6/219 (2%)
Frame = +3
Query: 457 ISGGGSLPLEVDKFFEAI--GVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFK 514
+ GG L E D+ F+A V V GYGLTE S V N +G+ G + + E K
Sbjct: 129 VCGGSPLRKETDEAFKAKFPNVLVMQGYGLTE-SAVTRTTPEEANQVGATGKLIPNIEAK 305
Query: 515 VVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHST 574
+V+ ETGE + PG +G L +RGP VM GY +P AT+ L DGWL TGDL +
Sbjct: 306 IVNPETGEAMFPGEQGELWIRGPYVMKGYSGDPKATSATL-VDGWLRTGDLCYF------ 464
Query: 575 GRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG--- 631
+S G + V R K+ ++ G V PAELEE + S I VI G
Sbjct: 465 ----DSKGFLYVVDRLKE-LIKYKGYQVAPAELEELLLSHSEINDAAVIPYPDEVAGQVP 629
Query: 632 -AIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIY 669
A +V + L A +ID ++ V +K+ + +
Sbjct: 630 MAFVVRQPQSSLGAA---EVIDFVAKQVSPYKKIRRVAF 737
>TC205641 UP|Q8S5C1 (Q8S5C1) 4-coumarate:CoA ligase isoenzyme 2 , complete
Length = 1946
Score = 98.6 bits (244), Expect = 9e-21
Identities = 120/461 (26%), Positives = 181/461 (39%), Gaps = 8/461 (1%)
Frame = +1
Query: 262 INSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWD----TVPAEVGDRFLSMLPPWH 317
I DD+ L Y+SGTTG PKGVML+H+ L+ I D + D L +LP +H
Sbjct: 610 IKPDDVVALPYSSGTTGLPKGVMLSHKGLVTSIAQQVDGDNPNLYYHCHDTILCVLPLFH 789
Query: 318 AYERACEYFIFTCGIEQVYTTVRNLKDD----LGRYQPHYMISVPLVFETLYSGIQKQIS 373
Y + CG+ T + K D L H + P+V I IS
Sbjct: 790 IYSLNS---VLLCGLRAKATILLMPKFDINSLLALIHKHKVTIAPVV-----PPIVLAIS 945
Query: 374 TSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFP 433
SP + K L+ IRV + G L + ++
Sbjct: 946 KSPDLHK-YDLSSIRVLKS--------GGAPLGKELED---------------------- 1032
Query: 434 IHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAA 493
L KF +K+ G+++A G L + + E I VK
Sbjct: 1033--TLRAKFPNAKLGQGYGMTEA----GPVLTMSLAFAKEPIDVKP--------------- 1149
Query: 494 RRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQA 553
G+ G V++ E K+VD ETG LP G + +RG +M GY + AT +
Sbjct: 1150--------GACGTVVRNAEMKIVDPETGHSLPRNQSGEICIRGDQIMKGYLNDGEATERT 1305
Query: 554 LDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMR 613
+DKDGWL+TGD+G+I + + +VD R K+ ++ G V PAELE +
Sbjct: 1306IDKDGWLHTGDIGYI---------DDDDELFIVD-RLKE-LIKYKGFQVAPAELEALLLT 1452
Query: 614 SSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELK 673
I A +VP +E ++ S +E+++ I K++
Sbjct: 1453HPKISD------------AAVVPMKDEAAGEVPVAFVVISNGYTDTTEDEIKQFISKQVV 1596
Query: 674 TWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAK 714
+ +N F ID +P+ KI R + AK
Sbjct: 1597FYKR-----------INRVFFIDAIPKSPSGKILRKDLRAK 1686
>TC211377 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (58%)
Length = 1248
Score = 97.8 bits (242), Expect = 1e-20
Identities = 114/458 (24%), Positives = 183/458 (39%), Gaps = 18/458 (3%)
Frame = +1
Query: 118 ILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYN 177
+ +A G R E++A FAD RW +A QG V S E L N
Sbjct: 1 VSSFASGXXSXXXRXXERVAXFADTRERWFIALQGCFRRNVTVVTMYSSLGKEALCHSLN 180
Query: 178 HSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVI 237
+E + E+ + + + R I + SD + A+ ++ FS +E
Sbjct: 181 ETEVTTVICGRKELKSLVNISGQLDSVKRVICMDDDIPSDASS-AQHGWKITTFSNVE-- 351
Query: 238 DLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNL 297
LGRE+ + + S D+A ++YTSG+TG PKGVM+TH N+L + ++
Sbjct: 352 RLGRENPVEA------------DLPLSADVAVIMYTSGSTGLPKGVMMTHGNVLATVSSV 495
Query: 298 WDTVPAEVG--DRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKD----------- 344
VP +G D +L+ LP H E E I G Y + L D
Sbjct: 496 MIIVP-NLGPKDVYLAYLPMAHILELVAENLIAAVGGCIGYGSPLTLTDTSNKIKKGKQG 672
Query: 345 DLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKC 404
D P M +VP + + + G+ K++++ + K L ++ Y R +
Sbjct: 673 DSTALMPTVMAAVPAILDRVRDGVLKKVNSKGGLSK---------KLFHLAYSRRLQA-- 819
Query: 405 LTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAI-GLSKAGISGGGSL 463
+N W A F V+ K+ + + G + + GG L
Sbjct: 820 ----------INGCWFGAWGLEKALWNF--------LVFKKVQAILGGRIRFILCGGAPL 945
Query: 464 PLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGE 522
+ +F +G + GYGLTET + +G VG PV + K++D G
Sbjct: 946 SGDTQRFINICLGAPIGQGYGLTETCAGGSFSDFDDTSVGRVGPPVPCSYIKLIDWPEGG 1125
Query: 523 VL---PPGSKGILKVRGPPVMNGYYKNPLATNQALDKD 557
P ++G + + GP V GY+KN T ++ D
Sbjct: 1126YSTSDSPMARGEIVIGGPNVTLGYFKNEEKTKESYKVD 1239
>TC206424 similar to UP|Q8LKS6 (Q8LKS6) Long chain acyl-CoA synthetase 6 ,
partial (30%)
Length = 1009
Score = 93.2 bits (230), Expect = 4e-19
Identities = 56/203 (27%), Positives = 99/203 (48%), Gaps = 5/203 (2%)
Frame = +1
Query: 526 PGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIV 585
P +G + VRGP V GY+K+ T +D+DGWL+TGD+G P G +
Sbjct: 55 PNPRGEICVRGPIVFRGYHKDEAQTRDVIDEDGWLHTGDIGTWLP----------GGRLK 204
Query: 586 VDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQD-KRRLGAIIVPNSEEVLKV 644
+ R K+ L+ GE + P ++E + + Q V G L A++ + + +
Sbjct: 205 IIDRKKNIFKLAQGEYIAPEKIENVYAKCKFVAQCFVYGDSLNASLVAVVSVDHDNLKAW 384
Query: 645 ARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQ----IGPILLVNEPFTIDNGLM 700
A I+ + + + ++ K + E+ E+ + + + LV EPFT++NGL+
Sbjct: 385 AASEGIMYNDLAQLCNDPKARAAVLAEMDAAGREAQLRGFEFVKAVTLVLEPFTLENGLL 564
Query: 701 TPTMKIRRDRVVAKYKEQIDDLY 723
TPT K++R + + + I D+Y
Sbjct: 565 TPTFKVKRPQAKEYFAKAISDMY 633
>TC216929 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial (51%)
Length = 1432
Score = 92.0 bits (227), Expect = 8e-19
Identities = 99/402 (24%), Positives = 168/402 (41%), Gaps = 18/402 (4%)
Frame = +1
Query: 340 RNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKR- 398
+ K D +P + +VP + + + G+ K++ + K +L + YKR
Sbjct: 49 KGTKGDATVLKPTLLTAVPAILDRIRDGVVKKVEQKGGLEK---------NLFHFAYKRR 201
Query: 399 --IYEGKCL-TRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLS-K 454
+G L TR +++ +W I V+ +I SA+G +
Sbjct: 202 LAAVKGSWLRTRGLEK---------LMWDTI---------------VFKQIRSALGGQLR 309
Query: 455 AGISGGGSLPLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEF 513
+ GG L + F +G + GYGLTET +G VG P+
Sbjct: 310 FMLCGGAPLSGDSQHFINICMGATIGQGYGLTETFAGAVFSEWDDYSVGRVGPPLPCCHI 489
Query: 514 KVVDSETGEVLP---PGSKGILKVRGPPVMNGYYKNPLATNQA--LDKDG--WLNTGDLG 566
K+V E G L P +G + V G V GY+KN T + +D+ G W TGD+G
Sbjct: 490 KLVSWEEGGYLTSDKPMPRGEIVVGGFSVTAGYFKNQEKTKEVFKVDEKGMRWFYTGDIG 669
Query: 567 WIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQD 626
P G + + R KD + L GE + ++E A + I+V
Sbjct: 670 QFHP----------DGCLEIIDRKKDIVKLQHGEYISLGKVEAALSSCDYVDNIMVYADP 819
Query: 627 KRRLGAIIVPNSEEVLKVARELSIID-----SISSNVVSEEKVLNLIYKELKTWMSESPF 681
+V S++ L+ + + ID + + + +VL I K K+ E
Sbjct: 820 FHNYCVALVVASQQSLEKWAQQAGIDYQDFPDLCNKPETVTEVLQSISKVAKSAKLEKTE 999
Query: 682 QIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
I L+ +P+T ++GL+T +KI+R+++ AK+K+ + LY
Sbjct: 1000IPAKIKLLPDPWTPESGLVTAALKIKREQLKAKFKDDLQKLY 1125
>TC216190 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
complete
Length = 1943
Score = 92.0 bits (227), Expect = 8e-19
Identities = 68/218 (31%), Positives = 109/218 (49%), Gaps = 9/218 (4%)
Frame = +3
Query: 427 IATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFEAIGVKVQN-----G 481
+A+ + PI L +K + + + +A ++G L E+ EA+ ++ + G
Sbjct: 873 VASFVPPIVLALVKSGETHRYDLSSI-RAVVTGAAPLGGELQ---EAVKARLPHATFGQG 1040
Query: 482 YGLTETSPVIA----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGP 537
YG+TE P+ A+ P G+ G V++ E K+VD+ETG+ LP G + +RG
Sbjct: 1041 YGMTEAGPLAISMAFAKEPSKIKPGACGTVVRNAEMKIVDTETGDSLPRNKSGEICIRGT 1220
Query: 538 PVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLS 597
VM GY +P AT + +DK+GWL+TGD+G+I + + +VD R K+ ++
Sbjct: 1221 KVMKGYLNDPEATERTIDKEGWLHTGDIGFI---------DDDDELFIVD-RLKE-LIKY 1367
Query: 598 TGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
G V PAELE + I V+G G I V
Sbjct: 1368 KGFQVAPAELEALLIAHPNISDAAVVGMKDEAAGEIPV 1481
Score = 56.6 bits (135), Expect = 4e-08
Identities = 61/247 (24%), Positives = 96/247 (38%), Gaps = 5/247 (2%)
Frame = +3
Query: 91 KYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVAD 150
K+ D+ L+D T+TY ++ A A GL IG+R + + L N ++ +A
Sbjct: 138 KFHDRPCLIDG--DTGETLTYADVDLAARRIASGLHKIGIRQGDVIMLVLRNCPQFALAF 311
Query: 151 QGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIIL 210
G GA+ V Y +E A + T R +I
Sbjct: 312 LGATHRGAV---------VTTANPFYTPAELAKQA---------------TATKTRLVIT 419
Query: 211 LWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEA-INSDDIAT 269
+ A+ + +V + + + + S A + IN D++
Sbjct: 420 QSAYVEKIKSFADSSSDVMVMCIDDDFSYENDGVLHFSTLSNADETEAPAVKINPDELVA 599
Query: 270 LIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVP----AEVGDRFLSMLPPWHAYERACEY 325
L ++SGT+G PKGVML+H+NL+ I L D D L +LP +H Y
Sbjct: 600 LPFSSGTSGLPKGVMLSHKNLVTTIAQLVDGENPHQYTHSEDVLLCVLPMFHIYALNS-- 773
Query: 326 FIFTCGI 332
I CGI
Sbjct: 774 -ILLCGI 791
>TC223899 similar to UP|Q8LRT1 (Q8LRT1) Acyl-CoA synthetase-like protein,
partial (7%)
Length = 428
Score = 89.7 bits (221), Expect = 4e-18
Identities = 42/58 (72%), Positives = 50/58 (85%)
Frame = +2
Query: 667 LIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
L + L SESPFQIGP+LLVN+PFTIDNGLMTPTMKIRRD+VVA+Y +QI++LYK
Sbjct: 191 LFFLSLSCRTSESPFQIGPVLLVNDPFTIDNGLMTPTMKIRRDKVVAQYGDQIENLYK 364
>TC227531 similar to UP|Q84P23 (Q84P23) 4-coumarate-CoA ligase-like protein,
partial (59%)
Length = 1478
Score = 88.6 bits (218), Expect = 9e-18
Identities = 65/182 (35%), Positives = 89/182 (48%), Gaps = 6/182 (3%)
Frame = +1
Query: 458 SGGGSLPLEVDKFFEAI--GVKVQNGYGLTET----SPVIAARRPRCNVIGSVGHPVQHT 511
SGG L EV + F A V++ GYGLTE+ + V+ + + GSVG ++
Sbjct: 445 SGGAPLGKEVAEDFRAQFPNVEIGQGYGLTESGGGAARVLGPDESKRH--GSVGRLSENM 618
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
E K+VD TGE L PG KG L +RGP +M GY + AT + LD +GWL TGDL +
Sbjct: 619 EAKIVDPVTGEALSPGQKGELWLRGPTIMKGYVGDEKATAETLDSEGWLKTGDLCYF--- 789
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
+S G + + R K+ I + V PAELE + I V+ G
Sbjct: 790 -------DSDGFLYIVDRLKELIKYKAYQ-VPPAELEHILHTNPEIADAAVVPYPDEEAG 945
Query: 632 AI 633
I
Sbjct: 946 QI 951
Score = 42.0 bits (97), Expect = 0.001
Identities = 37/128 (28%), Positives = 59/128 (45%), Gaps = 14/128 (10%)
Frame = +1
Query: 262 INSDDIATLIYTSGTTGNPKGVMLTHRNLL------HQIKNLWDTVPAEVGDRFLSMLPP 315
++ D A ++++SGTTG KGV+LTHRN + + ++ + D P V L LP
Sbjct: 64 VSQSDSAAILFSSGTTGRVKGVLLTHRNFIALIGGFYHLRMVVDDDPHPVS---LFTLPL 234
Query: 316 WHAYERACEYFIFTCGIEQVYTTV-------RNLKDDLGRYQPHYM-ISVPLVFETLYSG 367
+H + +F+ I T V + + RY+ YM +S PLV S
Sbjct: 235 FHVF----GFFMLVRAIAVGETLVFMHRFDFEGMLKAVERYRITYMPVSPPLVVALAKSE 402
Query: 368 IQKQISTS 375
+ K+ S
Sbjct: 403 LVKKYDMS 426
>TC229241 similar to UP|Q9M0X9 (Q9M0X9) 4-coumarate-CoA ligase-like protein,
partial (37%)
Length = 986
Score = 86.7 bits (213), Expect = 3e-17
Identities = 70/238 (29%), Positives = 110/238 (45%), Gaps = 2/238 (0%)
Frame = +3
Query: 484 LTETSPVIAARRPRCNV--IGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMN 541
+TET +++ PR V GS G V E ++V +T + LPP G + VRGP +M
Sbjct: 3 MTETCGIVSVENPRVGVRHTGSTGTLVSGVEAQIVSVDTQKPLPPRQLGEIWVRGPNMMQ 182
Query: 542 GYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGEN 601
GY+ NP AT +DK GW++TGDLG+ + G + V R K+ ++ G
Sbjct: 183 GYHNNPEATRLTIDKKGWVHTGDLGYF----------DEDGQLYVVDRIKE-LIKYKGFQ 329
Query: 602 VEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSE 661
V PAELE + I L A++VP ++ ++ S +S+ ++E
Sbjct: 330 VAPAELEGLLVSHPEI------------LEAVVVPYPDDEAGEVPIAYVVRSPNSS-LTE 470
Query: 662 EKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQI 719
E++ I K Q+ P + I+N T + KI R + AK + +I
Sbjct: 471 EEIQKFIAK-----------QVAPFKKLRRVTFINNVPKTASGKILRRELTAKARSKI 611
>TC232071 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (35%)
Length = 962
Score = 85.5 bits (210), Expect = 8e-17
Identities = 69/218 (31%), Positives = 103/218 (46%), Gaps = 9/218 (4%)
Frame = +2
Query: 461 GSLPL--EVDKFFEAI--GVKVQNGYGLTETSPVIAARRPR-CNVIGSVGHPVQHTEFKV 515
GS PL E + F+A V + GYGLTE++ +A P N G+ G V E K+
Sbjct: 71 GSSPLGKETAEAFKAKFPNVMILQGYGLTESTAGVARTSPEDANRAGTTGRLVSGVEAKI 250
Query: 516 VDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTG 575
V+ TGE + P +G L ++ P +M GY +P AT+ L DGWL TGDL +
Sbjct: 251 VNPNTGEAMFPCEQGELWIKSPSIMKGYVGDPEATSATL-VDGWLRTGDLCYF------- 406
Query: 576 RSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG---- 631
++ G + V R K+ ++ G V PAELE+ + I VI G
Sbjct: 407 ---DNEGFLYVVDRLKE-LIKYKGYQVAPAELEQYLLSHPEINDAAVIPYPDEEAGQVPM 574
Query: 632 AIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIY 669
A +V + L E+ IID ++ V +K+ + +
Sbjct: 575 AFVVRQPQSSLS---EIEIIDFVAKQVAPYKKIRRVAF 679
>TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL 2)
(4-coumaroyl-CoA synthase 2) (Clone 4CL16) , complete
Length = 1973
Score = 83.2 bits (204), Expect = 4e-16
Identities = 54/160 (33%), Positives = 80/160 (49%), Gaps = 5/160 (3%)
Frame = +1
Query: 481 GYGLTETSPVIA-----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVR 535
GYG+TE PV++ A++P GS G V++ E KVVD ETG L G + +R
Sbjct: 1081 GYGMTEAGPVLSMCLGFAKQPFQTKSGSCGTVVRNAELKVVDPETGRSLGYNQPGEICIR 1260
Query: 536 GPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIV 595
G +M GY + AT +D +GWL+TGD+G++ + + +VD R K+ ++
Sbjct: 1261 GQQIMKGYLNDEAATASTIDSEGWLHTGDVGYV---------DDDDEIFIVD-RVKE-LI 1407
Query: 596 LSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
G V PAELE + I V+ Q G + V
Sbjct: 1408 KYKGFQVPPAELEGLLVSHPSIADAAVVPQKDVAAGEVPV 1527
Score = 42.0 bits (97), Expect = 0.001
Identities = 29/88 (32%), Positives = 46/88 (51%), Gaps = 5/88 (5%)
Frame = +1
Query: 237 IDLGRESRMALSDSHEASQRYVYEA-INSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIK 295
+D E+ + S EA++ V E I+ DD + ++SGTTG PKGV+LTH++L +
Sbjct: 541 VDDPPENCLHFSVLSEANESDVPEVEIHPDDAVAMPFSSGTTGLPKGVILTHKSLTTSVA 720
Query: 296 NLWD----TVPAEVGDRFLSMLPPWHAY 319
D + D L +LP +H +
Sbjct: 721 QQVDGENPNLYLTTEDVLLCVLPLFHIF 804
>TC210966 similar to UP|Q96338 (Q96338) AMP-binding protein , partial (32%)
Length = 648
Score = 82.4 bits (202), Expect = 6e-16
Identities = 67/235 (28%), Positives = 107/235 (45%), Gaps = 7/235 (2%)
Frame = +3
Query: 265 DDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVG--DRFLSMLPPWHAYERA 322
DDIAT+ YTSGTTG PKG +LTH N + + T+ + G D ++S LP H YERA
Sbjct: 39 DDIATICYTSGTTGTPKGAILTHGNFIASVAG--STMDEKFGPSDVYISYLPLAHIYERA 212
Query: 323 CEYFIFTCGIEQVYTTVRNLK--DDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRK 380
+ GI + ++K DD+ +P SVP ++ +Y+GI + TS +++
Sbjct: 213 NQVMTVHFGIAVGFYQGDSMKLMDDIAALRPTVFCSVPRLYNRIYAGITNAVKTSGGLKE 392
Query: 381 LVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIK 440
+ + Y K +Q + +W R+
Sbjct: 393 RL-------------FNAAYNAK------RQALLHGKNPSPMWDRL-------------- 473
Query: 441 FVYSKIHSAIGLSKAGISGGGSLPLEVD--KFFE-AIGVKVQNGYGLTETSPVIA 492
V++KI +G + G+ PL D +F + G +V GYG+TE++ VI+
Sbjct: 474 -VFNKIKEKLG-GRVRFMASGASPLSPDIMEFLKICFGCRVTEGYGMTESTCVIS 632
>TC217745 similar to UP|Q96538 (Q96538) Acyl-CoA synthetase , partial (37%)
Length = 986
Score = 81.6 bits (200), Expect = 1e-15
Identities = 70/254 (27%), Positives = 116/254 (45%), Gaps = 7/254 (2%)
Frame = +1
Query: 478 VQNGYGLTETSPVIAARRPR-CNVIGSVGHPVQHTE--FKVVDSETGEVLPPGSKGILKV 534
V GYGLTET P ++G+VG PV + + + V + L +G + V
Sbjct: 16 VLQGYGLTETCAGTFVSLPNEIEMLGTVGPPVPNVDVCLESVPEMGYDALASTPRGEICV 195
Query: 535 RGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTI 594
+G + GYYK T + L D W +TGD+G P+ G + + R K+
Sbjct: 196 KGKTLFAGYYKREDLTKEVLI-DEWFHTGDIGEWQPN----------GSMKIIDRKKNIF 342
Query: 595 VLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSI 654
LS GE V LE + S I+ I V G +V S++ L+ + + I
Sbjct: 343 KLSQGEYVAVENLENIYGQVSSIESIWVYGNSFEAFLVAVVNPSKQALEHWAQENGISMD 522
Query: 655 SSNVVSEEKVLNLIYKELKTWMSESPFQ----IGPILLVNEPFTIDNGLMTPTMKIRRDR 710
+++ + + + I +EL E + I + L + PF ++ L+TPT K +R +
Sbjct: 523 FNSLCEDARAKSYIIEELSKIAKEKKLKGFEFIKAVHLDSIPFDMERDLITPTYKKKRPQ 702
Query: 711 VVAKYKEQIDDLYK 724
++ Y+ ID++YK
Sbjct: 703 LLKYYQNAIDNMYK 744
>TC224144 homologue to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (38%)
Length = 783
Score = 80.5 bits (197), Expect = 2e-15
Identities = 38/92 (41%), Positives = 58/92 (62%), Gaps = 4/92 (4%)
Frame = +2
Query: 481 GYGLTETSPVIA----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRG 536
GYG+TE P+ A+ P G+ G V++ E K+VD+ETG+ LP G + +RG
Sbjct: 326 GYGMTEAGPLAISMAFAKEPSKIKPGACGTVVRNAEMKIVDTETGDSLPRNKSGEICIRG 505
Query: 537 PPVMNGYYKNPLATNQALDKDGWLNTGDLGWI 568
VM GY +P AT + +D++GW++TGD+G+I
Sbjct: 506 AKVMKGYLNDPEATERTIDREGWVHTGDIGFI 601
>TC226637 UP|Q8S5C2 (Q8S5C2) 4-coumarate:CoA ligase isoenzyme 3 , complete
Length = 1934
Score = 80.5 bits (197), Expect = 2e-15
Identities = 53/160 (33%), Positives = 79/160 (49%), Gaps = 5/160 (3%)
Frame = +1
Query: 481 GYGLTETSPVIA-----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVR 535
GYG+TE PV++ A++P GS G V++ E +VVD ETG L G + +R
Sbjct: 1072 GYGMTEAGPVLSMCLGFAKQPFPTKSGSCGTVVRNAELRVVDPETGRSLGYNQPGEICIR 1251
Query: 536 GPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIV 595
G +M GY + AT +D +GWL+TGD+G++ + +VD R K+ ++
Sbjct: 1252 GQQIMKGYLNDEKATALTIDSEGWLHTGDVGYV---------DEDDEIFIVD-RVKE-LI 1398
Query: 596 LSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIV 635
G V PAELE + I V+ Q G + V
Sbjct: 1399 KYKGFQVPPAELEGLLVSHPSIADAAVVPQKDVAAGEVPV 1518
Score = 42.0 bits (97), Expect = 0.001
Identities = 45/217 (20%), Positives = 83/217 (37%), Gaps = 5/217 (2%)
Frame = +1
Query: 108 TITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRS 167
T TY++ A GL +G+R + + + NS ++ + GA+
Sbjct: 220 TYTYSETHLISRKIAAGLSNLGIRKGDVVMILLQNSAEFVFSFLAASMIGAVATTANPFY 399
Query: 168 SVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKE 227
+ E+ + + S++ + + +K G F ++
Sbjct: 400 TAAEIFKQFTVSKTKLIITQAMYVDKLRNHDDGAKLGEDFKVV----------------- 528
Query: 228 VPIFSFMEVIDLGRESRMALSDSHEASQRYVYEA-INSDDIATLIYTSGTTGNPKGVMLT 286
+D E+ + S EA++ E I DD + ++SGTTG PKGV+LT
Sbjct: 529 --------TVDDPPENCLHFSVLSEANESDAPEVDIQPDDAVAMPFSSGTTGLPKGVVLT 684
Query: 287 HRNLLHQIKNLWD----TVPAEVGDRFLSMLPPWHAY 319
H++L + D + D L +LP +H +
Sbjct: 685 HKSLTTSVAQQVDGENPNLYLTTEDVLLCVLPLFHIF 795
>TC216082 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (40%)
Length = 909
Score = 80.1 bits (196), Expect = 3e-15
Identities = 60/199 (30%), Positives = 92/199 (46%), Gaps = 5/199 (2%)
Frame = +1
Query: 476 VKVQNGYGLTETSPVIAARRP--RCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILK 533
V + GYGLTE++ V A+ G+ G T+ +VD E+G+ LP G L
Sbjct: 34 VTILQGYGLTESTGVGASTDSLEESRRYGTAGLLSPATQAMIVDPESGQSLPVNRTGELW 213
Query: 534 VRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDT 593
+RGP +M GY+ N AT LD GWL TGD+ +I ++ G I + R K+
Sbjct: 214 LRGPTIMKGYFSNEEATTSTLDSKGWLRTGDICYI----------DNDGFIFIVDRLKE- 360
Query: 594 IVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELS---I 650
++ G V PAELE + I VI + G P + V K LS +
Sbjct: 361 LIKYKGYQVPPAELEALLLTHPAILDAAVIPYPDKEAGQ--HPMAYVVRKAGSSLSETQV 534
Query: 651 IDSISSNVVSEEKVLNLIY 669
+D ++ V +++ + +
Sbjct: 535 MDFVAGQVAPYKRIRKVAF 591
>TC227478 similar to UP|Q96537 (Q96537) Acyl CoA synthetase , partial (28%)
Length = 972
Score = 79.7 bits (195), Expect = 4e-15
Identities = 56/201 (27%), Positives = 100/201 (48%), Gaps = 5/201 (2%)
Frame = +3
Query: 529 KGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLG-WIAPHHSTGRSRNSSGVIVVD 587
+G + +RG + +GY+K T + + DGW +TGD+G W S+G + +
Sbjct: 3 RGEICLRGNTLFSGYHKREDLTKEVM-VDGWFHTGDIGEW-----------QSNGAMKII 146
Query: 588 GRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQD-KRRLGAIIVPNSEEVLKVAR 646
R K+ LS GE + +E ++ +I I V G + L A++VP + + A+
Sbjct: 147 DRKKNIFKLSQGEYIAVENIENKYLQCPLIASIWVYGNSFESFLVAVVVPERKAIEDWAK 326
Query: 647 ELSIID---SISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPT 703
E ++ D S+ N+ + + +L+ + + + I L PF I+ L+TPT
Sbjct: 327 EHNLTDDFKSLCDNLKARKHILDELNSTGQKHQLRGFELLKAIHLEPNPFDIERDLITPT 506
Query: 704 MKIRRDRVVAKYKEQIDDLYK 724
K++R +++ YK+ ID LYK
Sbjct: 507 FKLKRPQLLKYYKDHIDQLYK 569
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,945,358
Number of Sequences: 63676
Number of extensions: 425530
Number of successful extensions: 2935
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 2412
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2755
length of query: 724
length of database: 12,639,632
effective HSP length: 104
effective length of query: 620
effective length of database: 6,017,328
effective search space: 3730743360
effective search space used: 3730743360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146585.9


