

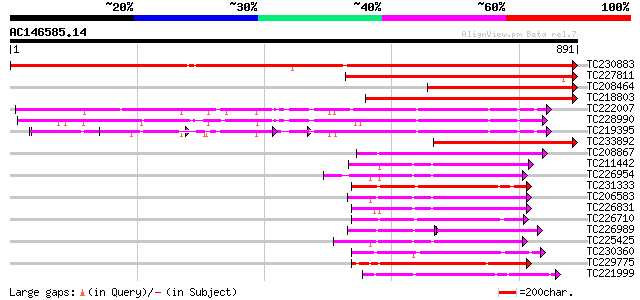
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.14 - phase: 0
(891 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 1168 0.0
TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , rece... 476 e-134
TC208464 homologue to UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RH... 395 e-110
TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kin... 385 e-107
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 316 3e-86
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 305 6e-83
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 300 2e-81
TC233892 similar to UP|Q84P72 (Q84P72) Receptor-like protein kin... 239 4e-63
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 229 3e-60
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 222 5e-58
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 218 1e-56
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 217 2e-56
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 217 2e-56
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 209 6e-54
TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-l... 206 3e-53
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 126 7e-53
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 204 1e-52
TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {A... 204 2e-52
TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 202 7e-52
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 201 9e-52
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 1168 bits (3021), Expect = 0.0
Identities = 601/902 (66%), Positives = 695/902 (76%), Gaps = 11/902 (1%)
Frame = +1
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
M + +SL P P+GWS T FCQW G++C S + VTSI+L+ Q L GTLP +LNSL+QL
Sbjct: 1 MSNFLKSLTPPPSGWSETTPFCQWKGIQCDSSSHVTSISLASQSLTGTLPSDLNSLSQLR 180
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
TL LQ+N+L+G LPSL+NLS L V NNFSSV+P AF+ L SLQTLSLG N L PW
Sbjct: 181 TLSLQDNSLTGTLPSLSNLSFLQTVYFNRNNFSSVSPTAFASLTSLQTLSLGSNPALQPW 360
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSL-AGSGI 179
+FPT+LT SSNL +D+ + G LPDIF F SL L L+YNNL+G LP+S A + +
Sbjct: 361 SFPTDLTSSSNLIDLDLATVSLTGPLPDIFDKFPSLQHLRLSYNNLTGNLPSSFSAANNL 540
Query: 180 QSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTG 239
++ W+NN GL+G++ V+SNM+ L Q WL+ N+FTG IPDLSQC ++ DLQLRDNQLTG
Sbjct: 541 ETLWLNNQAAGLSGTLLVLSNMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLTG 720
Query: 240 VVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMD 299
VVP SL S+ L+ V+L NN+LQGPVPVFGK V D I N+FC + CD RVM
Sbjct: 721 VVPASLTSLPSLKKVSLDNNELQGPVPVFGKGVNVTLDGI--NSFCLDTPG-NCDPRVMV 891
Query: 300 LLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTD 359
LL I FGYPI+ A+SW GNDPC W V+C GKI +NF KQGLQGTISPAFANLTD
Sbjct: 892 LLQIAEAFGYPIRLAESWKGNDPCDGWNYVVCAAGKIITVNFEKQGLQGTISPAFANLTD 1071
Query: 360 LTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGK-- 417
L L+LNGNNL GSIP +L TL QL+TLDVS+N+LSG VPKF PKVK +T GN LGK
Sbjct: 1072LRTLFLNGNNLIGSIPDSLITLPQLQTLDVSDNNLSGLVPKFPPKVKLVTAGNALLGKPL 1251
Query: 418 NHGGGAPGSAPGGSPAGSGKGASMK------KVWIIIIIVLIVVGFVVGGAWFSWKCYSR 471
+ GGG G+ P GS G G S K WI I+V IV+ F+ + SWKC+
Sbjct: 1252SPGGGPSGTTPSGSSTGGSGGESSKGNSSVSPGWIAGIVV-IVLFFIAVVLFVSWKCFVN 1428
Query: 472 KGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNAT 531
K +F+RV ENG+G KLD VSNGYGG ELQSQSSGD SDLH DG T
Sbjct: 1429KLQGKFSRVKGHENGKGGFKLDAVHVSNGYGGVPVELQSQSSGDRSDLHALDG-----PT 1593
Query: 532 ISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEI 591
SI VL+QVTN+FS++NILGRGGFG+VYKG+L DGTKIAVKRM SVA G+KGL EF+AEI
Sbjct: 1594FSIQVLQQVTNNFSEENILGRGGFGVVYKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEI 1773
Query: 592 GVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIA 651
VL+KVRHRHLVALLGYCING ERLLVYE+MPQGTLTQHLFE +E GY PLTWKQR++IA
Sbjct: 1774AVLSKVRHRHLVALLGYCINGIERLLVYEYMPQGTLTQHLFEWQEQGYVPLTWKQRVVIA 1953
Query: 652 LDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAG 711
LDV RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG YSVET+LAG
Sbjct: 1954LDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAG 2133
Query: 712 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNK 771
TFGYLAPEYAATGRVTTKVD+YAFG+VLMELITGRKALDD+VPDE SHLVTWFRRVL NK
Sbjct: 2134TFGYLAPEYAATGRVTTKVDIYAFGIVLMELITGRKALDDTVPDERSHLVTWFRRVLINK 2313
Query: 772 ENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTT 831
ENIPKAIDQTL+PDEETM SIYKVAELAGHCT R PYQRPD+GHAVNVL PLV+QW+P++
Sbjct: 2314ENIPKAIDQTLNPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSS 2493
Query: 832 HTDEST-CADDNQMSLTQALQRWQANEGTSTFFNGMT-SQTQSSSTSKPPVFADSLHSPD 889
H +E D QMSL QAL+RWQANEGTS+ FN ++ SQTQSS +SKP FAD+ S D
Sbjct: 2494HDEEEDGSGGDLQMSLPQALRRWQANEGTSSIFNDISISQTQSSISSKPVGFADTFDSMD 2673
Query: 890 CR 891
CR
Sbjct: 2674CR 2679
>TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , receptor type
precursor - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (40%)
Length = 1436
Score = 476 bits (1225), Expect = e-134
Identities = 239/371 (64%), Positives = 289/371 (77%), Gaps = 7/371 (1%)
Frame = +2
Query: 528 GNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEF 587
GN ISI VL+ VT++FS+ N+LG+GGFG VY+GEL DGT+IAVKRM A KG EF
Sbjct: 62 GNMVISIQVLKNVTDNFSEKNVLGQGGFGTVYRGELHDGTRIAVKRMECGAIAGKGAAEF 241
Query: 588 QAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQR 647
++EI VLTKVRHRHLV+LLGYC++GNE+LLVYE+MPQGTL++HLF+ E G PL W +R
Sbjct: 242 KSEIAVLTKVRHRHLVSLLGYCLDGNEKLLVYEYMPQGTLSRHLFDWPEEGLEPLEWNRR 421
Query: 648 LIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVET 707
L IALDV RGVEYLH LA QSFIHRDLKPSNILLGDDMRAKVADFGLV+ AP+G S+ET
Sbjct: 422 LTIALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKASIET 601
Query: 708 KLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRV 767
++AGTFGYLAPEYA TGRVTTKVDV++FGV+LMELITGRKALD++ P++S HLVTWFRR+
Sbjct: 602 RIAGTFGYLAPEYAVTGRVTTKVDVFSFGVILMELITGRKALDETQPEDSMHLVTWFRRM 781
Query: 768 LTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQW 827
NK++ KAID T++ +EET+ SI+ VAELAGHC R PYQRPD+GHAVNVL LV+ W
Sbjct: 782 SINKDSFRKAIDSTIELNEETLASIHTVAELAGHCGAREPYQRPDMGHAVNVLSSLVELW 961
Query: 828 EPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS-------QTQSSSTSKPPV 880
+P+ E D MSL QAL++WQA EG S + +S TQ+S ++P
Sbjct: 962 KPSDQNSEDIYGIDLDMSLPQALKKWQAYEGRSQMESSASSSLLPSLDNTQTSIPTRPYG 1141
Query: 881 FADSLHSPDCR 891
FADS S D R
Sbjct: 1142FADSFTSADGR 1174
>TC208464 homologue to UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, partial
(26%)
Length = 893
Score = 395 bits (1015), Expect = e-110
Identities = 197/235 (83%), Positives = 208/235 (87%)
Frame = +2
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYL 716
GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG YSVET+LAGTFGYL
Sbjct: 2 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL 181
Query: 717 APEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPK 776
APEYAATGRVTTKVDVYAFGVVLMELITGR+ALDD+VPDE SHLV+WFRRVL NKENIPK
Sbjct: 182 APEYAATGRVTTKVDVYAFGVVLMELITGRRALDDTVPDERSHLVSWFRRVLINKENIPK 361
Query: 777 AIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDES 836
AIDQTLDPDEETM SIYKVAELAGHCT R PYQRPD+GHAVNVL PLV+QW+PTTH +E
Sbjct: 362 AIDQTLDPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLGPLVEQWKPTTHEEEE 541
Query: 837 TCADDNQMSLTQALQRWQANEGTSTFFNGMTSQTQSSSTSKPPVFADSLHSPDCR 891
D MSL QAL+RWQANEGTST F+ SQTQSS +KP FADS S DCR
Sbjct: 542 GYGIDLHMSLPQALRRWQANEGTSTMFDMSISQTQSSIPAKPSGFADSFDSMDCR 706
>TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kinase-like
protein (Fragment), partial (35%)
Length = 1266
Score = 385 bits (990), Expect = e-107
Identities = 196/334 (58%), Positives = 244/334 (72%), Gaps = 1/334 (0%)
Frame = +1
Query: 559 YKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLV 618
++GEL DGTKIAVKRM + SK L+EFQ+EI VL+KVRHRHLV+LLGY GNER+LV
Sbjct: 7 FRGELDDGTKIAVKRMEAGVISSKALDEFQSEIAVLSKVRHRHLVSLLGYSTEGNERILV 186
Query: 619 YEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSN 678
YE+MPQG L++HLF + H PL+WK+RL IALDV RG+EYLH+LA QSFIHRDLKPSN
Sbjct: 187 YEYMPQGALSKHLFHWKSHDLEPLSWKRRLNIALDVARGMEYLHTLAHQSFIHRDLKPSN 366
Query: 679 ILLGDDMRAKVADFGLVKNAPDG-NYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGV 737
ILL DD +AKV+DFGLVK AP+G SV T+LAGTFGYLAPEYA TG++TTK DV++FGV
Sbjct: 367 ILLADDFKAKVSDFGLVKLAPEGEKASVVTRLAGTFGYLAPEYAVTGKITTKADVFSFGV 546
Query: 738 VLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAE 797
VLMEL+TG ALD+ P+ES +L WF + ++K+ + AID LD EET S+ +AE
Sbjct: 547 VLMELLTGLMALDEDRPEESQYLAAWFWHIKSDKKKLMAAIDPALDVKEETFESVSIIAE 726
Query: 798 LAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANE 857
LAGHCT R P QRPD+GHAVNVL PLV++W+P E D + L Q ++ WQ E
Sbjct: 727 LAGHCTAREPSQRPDMGHAVNVLAPLVEKWKPFDDDTEEYSGIDYSLPLNQMVKGWQEAE 906
Query: 858 GTSTFFNGMTSQTQSSSTSKPPVFADSLHSPDCR 891
G + + ++SS ++P FADS S D R
Sbjct: 907 GKDLSYMDL-EDSKSSIPARPTGFADSFTSADGR 1005
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 316 bits (809), Expect = 3e-86
Identities = 273/985 (27%), Positives = 437/985 (43%), Gaps = 143/985 (14%)
Frame = +3
Query: 10 PSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNAL 69
P + W+++ +C W GV C + VT++NL+ L+GTL ++ L L+ L L N
Sbjct: 147 PVLSSWNASIPYCSWLGVTCDNRRHVTALNLTGLDLSGTLSADVAHLPFLSNLSLAANKF 326
Query: 70 SGPLP-SLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENI------------- 115
SGP+P SL+ LS L +NL +N F+ P L SL+ L L N
Sbjct: 327 SGPIPPSLSALSGLRYLNLSNNVFNETFPSELWRLQSLEVLDLYNNNMTGVLPLAVAQMQ 506
Query: 116 ---------NLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAY-NN 165
N P E + L + ++ +++GT+P G+ +SL L++ Y N
Sbjct: 507 NLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNELDGTIPPEIGNLTSLRELYIGYYNT 686
Query: 166 LSGGLPNSLAGSGIQSFWINNNLPGLTGSI-TVISNMTLLTQVWLHVNKFTGPI-PDLSQ 223
+GG+P + G+ + ++ L+G I + + L ++L VN +G + P+L
Sbjct: 687 YTGGIPPEI-GNLSELVRLDVAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGN 863
Query: 224 CNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVF--------------- 268
S+K + L +N L+G +P S + + + L N+L G +P F
Sbjct: 864 LKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWEN 1043
Query: 269 ----------GKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYP-------- 310
GK+ + N D+S N C + L +G F +
Sbjct: 1044 NLTGSIPEGLGKNGRLNLVDLSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESLGT 1223
Query: 311 ------IQFAKSWTGNDPCKDWLCVICGGGKITK------------------------LN 340
I+ +++ K + G K+T+ +
Sbjct: 1224 CESLTRIRMGENFLNGSIPKG----LFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQIT 1391
Query: 341 FAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLET-------------- 386
+ L G +SP+ N + + L L+GN TG IP + L QL
Sbjct: 1392 LSNNQLSGALSPSIGNFSSVQKLLLDGNMFTGRIPTQIGRLQQLSKIDFSGNKFSGPIAP 1571
Query: 387 ----------LDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSAPGGSPAGSG 436
LD+S N+LSG++P ++ + N L KNH GS P
Sbjct: 1572 EISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLN--LSKNH---LVGSIPSSI----- 1721
Query: 437 KGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLAS 496
+SM+ + + + G V G FS+ Y+ + +GNP+ +
Sbjct: 1722 --SSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYT-------SFLGNPDLCGPYLGACKGG 1874
Query: 497 VSNG-----YGGASSELQ---------------------SQSSGDHSDLHGFDGGNGGNA 530
V+NG G SS L+ ++S S+ +
Sbjct: 1875 VANGAHQPHVKGLSSSLKLLLVVGLLLCSIAFAVAAIFKARSLKKASEARAWKLTAFQRL 2054
Query: 531 TISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAE 590
++ V + +DNI+G+GG GIVYKG +P+G +AVKR+ ++++GS + F AE
Sbjct: 2055 DFTVD---DVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSHDHGFNAE 2225
Query: 591 IGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLII 650
I L ++RHRH+V LLG+C N LLVYE+MP G+L + + ++ G+ L W R I
Sbjct: 2226 IQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGE-VLHGKKGGH--LHWDTRYKI 2396
Query: 651 ALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GNYSVETKL 709
A++ +G+ YLH +HRD+K +NILL + A VADFGL K D G + +
Sbjct: 2397 AVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAI 2576
Query: 710 AGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVL- 768
AG++GY+APEYA T +V K DVY+FGVVL+ELITGRK + + + +V W R++
Sbjct: 2577 AGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEF--GDGVDIVQWVRKMTD 2750
Query: 769 TNKENIPKAIDQTLD--PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ 826
+NKE + K +D L P E M ++ VA L C +RP + V +L L
Sbjct: 2751 SNKEGVLKVLDPRLPSVPLHEVM-HVFYVAML---CVEEQAVERPTMREVVQILTELP-- 2912
Query: 827 WEPTTHTDESTCADDNQMSLTQALQ 851
+P + ++ +S + AL+
Sbjct: 2913 -KPPGSKEGDLTITESSLSSSNALE 2984
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 305 bits (781), Expect = 6e-83
Identities = 286/989 (28%), Positives = 423/989 (41%), Gaps = 157/989 (15%)
Frame = +1
Query: 13 NGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
+ W+S+T FC W G+ C S VTS+NL+ L+GTL D+L+ L L+ L L +N SGP
Sbjct: 223 SSWNSSTPFCSWFGLTCDSRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGP 402
Query: 73 LPS----------------------------LANLSSLTDVN------------------ 86
+P+ LANL L N
Sbjct: 403 IPASFSALSALRFLNLSNNVFNATFPSQLNRLANLEVLDLYNNNMTGELPLSVAAMPLLR 582
Query: 87 ---LGSNNFSSVTPGAFSGLNSLQTLSLGEN-----------------------INLSPW 120
LG N FS P + LQ L+L N N
Sbjct: 583 HLHLGGNFFSGQIPPEYGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIGYYNTYSG 762
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAG-SGI 179
P E+ SNL +D ++G +P G +L+TL L N LSG L L +
Sbjct: 763 GIPPEIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKSL 942
Query: 180 QSFWINNNLPG--LTGSITVISNMTLLT-----------------------QVWLHVNKF 214
+S ++NN+ + S + N+TLL Q+W N F
Sbjct: 943 KSMDLSNNMLSGEVPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLW--ENNF 1116
Query: 215 TGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVP-VFGKDV 272
TG IP +L + + L N++TG +P ++ + LQ + N L GP+P GK
Sbjct: 1117 TGSIPQNLGNNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCK 1296
Query: 273 KYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYP----IQFAKSW-TGNDPCKDWL 327
N + N N S+P G FG P ++ + TG P +
Sbjct: 1297 SLNRIRMGENFL---NGSIP-----------KGLFGLPKLTQVELQDNLLTGQFPEDGSI 1434
Query: 328 CVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETL 387
G +++ + L G++ N T + L LNGN TG IP + L QL +
Sbjct: 1435 ATDLG-----QISLSNNQLSGSLPSTIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKI 1599
Query: 388 D------------------------VSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGA 423
D +S N+LSGE+P ++ + N L +NH
Sbjct: 1600 DFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKITSMRILNYLN--LSRNH---L 1764
Query: 424 PGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNP 483
GS PG ASM+ + + G V G F + Y+ + +GNP
Sbjct: 1765 DGSIPGNI-------ASMQSLTSVDFSYNNFSGLVPGTGQFGYFNYT-------SFLGNP 1902
Query: 484 ENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTN- 542
E + V+NG + SS + G + A +I R +
Sbjct: 1903 ELCGPYLGPCKDGVANGPRQPHVKGPFSSSLKLLLVIGLLVCSILFAVAAIFKARALKKA 2082
Query: 543 --------------DFSDDNIL---------GRGGFGIVYKGELPDGTKIAVKRMISVAK 579
DF+ D++L G+GG GIVYKG +P+G +AVKR+ ++++
Sbjct: 2083 SEARAWKLTAFQRLDFTVDDVLDCLKEDNIIGKGGAGIVYKGAMPNGDNVAVKRLPAMSR 2262
Query: 580 GSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGY 639
GS + F AEI L ++RHRH+V LLG+C N LLVYE+MP G+L + + ++ G+
Sbjct: 2263 GSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGE-VLHGKKGGH 2439
Query: 640 TPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP 699
L W R IA++ +G+ YLH +HRD+K +NILL + A VADFGL K
Sbjct: 2440 --LHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNFEAHVADFGLAKFLQ 2613
Query: 700 D-GNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESS 758
D G + +AG++GY+APEYA T +V K DVY+FGVVL+EL+TGRK + + +
Sbjct: 2614 DSGASECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELVTGRKPVGEF--GDGV 2787
Query: 759 HLVTWFRRVL-TNKENIPKAIDQTLD--PDEETMLSIYKVAELAGHCTTRSPYQRPDIGH 815
+V W R++ +NKE + K +D L P E M ++ VA L C +RP +
Sbjct: 2788 DIVQWVRKMTDSNKEGVLKVLDSRLPSVPLHEVM-HVFYVAML---CVEEQAVERPTMRE 2955
Query: 816 AVNVLCPLVQQWEPTTHTDESTCADDNQM 844
V +L L + ES+ + N +
Sbjct: 2956 VVQILTELPKPPSSKHAITESSLSSSNSL 3042
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 300 bits (767), Expect = 2e-81
Identities = 258/932 (27%), Positives = 420/932 (44%), Gaps = 115/932 (12%)
Frame = +3
Query: 35 VTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPS-LANLSSLTDVNLGSNNFS 93
+++++L+ K +G +P +L++L+ L L L NN + PS L+ L +L ++L +NN +
Sbjct: 45 LSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRLQNLEVLDLYNNNMT 224
Query: 94 SVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGSF 153
V P A + + +L+ L LG N P E + L + ++ ++ GT+P G+
Sbjct: 225 GVLPLAVAQMQNLRHLHLGGNFFSGQ--IPPEYGRWQRLQYLAVSGNELEGTIPPEIGNL 398
Query: 154 SSLNTLHLAY-NNLSGGLPNSLAGSGIQSFWINNNLPGLTGSI-TVISNMTLLTQVWLHV 211
SSL L++ Y N +GG+P + G+ + ++ GL+G I + + L ++L V
Sbjct: 399 SSLRELYIGYYNTYTGGIPPEI-GNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQV 575
Query: 212 NKFTGPI-PDLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVF-- 268
N +G + P+L S+K + L +N L+G +P + + + L N+L G +P F
Sbjct: 576 NALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIG 755
Query: 269 -----------------------GKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVG 305
GK+ + N D+S N + C + L +G
Sbjct: 756 ELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLG 935
Query: 306 GF----------------------------------GYPIQFAKSWTGNDPCKDWLCVIC 331
F G P N ++ V
Sbjct: 936 NFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPEVGS 1115
Query: 332 GGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLET----- 386
+ ++ + L G + P+ N + + L L+GN TG IP + L QL
Sbjct: 1116VAVNLGQITLSNNQLSGVLPPSIGNFSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDFSG 1295
Query: 387 -------------------LDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNH-GGGAPGS 426
LD+S N+LSG++P ++ + N L +NH GG P S
Sbjct: 1296NKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLN--LSRNHLVGGIPSS 1469
Query: 427 APGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENG 486
+SM+ + + + G V G FS+ Y+ + +GNP+
Sbjct: 1470I-----------SSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYT-------SFLGNPDLC 1595
Query: 487 EGNVKLDLASVSNG-----YGGASSELQS---------------QSSGDHSDLHGFDGGN 526
+ V+NG G SS + + L G
Sbjct: 1596GPYLGACKDGVANGAHQPHVKGLSSSFKLLLVVGLLLCSIAFAVAAIFKARSLKKASGAR 1775
Query: 527 GGNATISIHV---LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKG 583
T + + V + +DNI+G+GG GIVYKG +P+G +AVKR+ ++++GS
Sbjct: 1776AWKLTAFQRLDFTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSH 1955
Query: 584 LNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLT 643
+ F AEI L ++RHRH+V LLG+C N LLVYE+MP G+L + + ++ G+ L
Sbjct: 1956DHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGE-VLHGKKGGH--LH 2126
Query: 644 WKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GN 702
W R IA++ +G+ YLH +HRD+K +NILL + A VADFGL K D G
Sbjct: 2127WDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGT 2306
Query: 703 YSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVT 762
+ +AG++GY+APEYA T +V K DVY+FGVVL+ELITGRK + + + +V
Sbjct: 2307SECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEF--GDGVDIVQ 2480
Query: 763 WFRRVL-TNKENIPKAIDQTLD--PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNV 819
W R++ +NKE + K +D L P E M ++ VA L C +RP + V +
Sbjct: 2481WVRKMTDSNKEGVLKVLDPRLPSVPLHEVM-HVFYVAML---CVEEQAVERPTMREVVQI 2648
Query: 820 LCPLVQQWEPTTHTDESTCADDNQMSLTQALQ 851
L L +P + + ++ +S + AL+
Sbjct: 2649LTELP---KPPDSKEGNLTITESSLSSSNALE 2735
Score = 116 bits (290), Expect = 5e-26
Identities = 124/486 (25%), Positives = 203/486 (41%), Gaps = 44/486 (9%)
Frame = +3
Query: 34 RVTSINLSDQKLAGTLPDNLNSLTQLTTLYL--QNNALSGPLPSLANLSSLTDVNLGSNN 91
R+ + +S +L GT+P + +L+ L LY+ N G P + NLS L ++
Sbjct: 330 RLQYLAVSGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCG 509
Query: 92 FSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFG 151
S P A L L TL L +N + EL +L S+D++ ++G +P FG
Sbjct: 510 LSGEIPAALGKLQKLDTLFL--QVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFG 683
Query: 152 SFSSLNTLHLAYNNLSGGLP---NSLAGSGIQSFWINN---NLP---------------- 189
++ L+L N L G +P L + W NN ++P
Sbjct: 684 ELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLSS 863
Query: 190 -GLTGSIT--VISNMTLLTQVWLHVNKFTGPIPD-LSQCNSIKDLQLRDNQLTGVVPDSL 245
LTG++ + S TL T + L N GPIP+ L C S+ +++ +N L G +P L
Sbjct: 864 NKLTGTLPTYLCSGNTLQTLITLG-NFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGL 1040
Query: 246 VSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIV- 304
+ L V L++N L G P G V N I+ +N + P + ++
Sbjct: 1041FGLPKLTQVELQDNYLSGEFPEVG-SVAVNLGQITLSNNQLSGVLPPSIGNFSSVQKLIL 1217
Query: 305 ----------GGFGYPIQFAK-SWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPA 353
G Q +K ++GN + I +T L+ ++ L G I
Sbjct: 1218DGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNE 1397
Query: 354 FANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVP---KFSPKVKFITD 410
+ L L L+ N+L G IP +++++ L ++D S N+LSG VP +FS
Sbjct: 1398ITGMRILNYLNLSRNHLVGGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFL 1577
Query: 411 GNVWLGKNHGGGAPGSAPGGSPAGSGKGASMK-KVWIIIIIVLIVVGFVVGGAWFSWKCY 469
GN L + G G+ KG S K+ +++ ++L + F V + +
Sbjct: 1578GNPDLCGPYLGACKDGVANGAHQPHVKGLSSSFKLLLVVGLLLCSIAFAVAAIFKARSLK 1757
Query: 470 SRKGLR 475
G R
Sbjct: 1758KASGAR 1775
Score = 105 bits (262), Expect = 9e-23
Identities = 79/255 (30%), Positives = 119/255 (45%), Gaps = 3/255 (1%)
Frame = +3
Query: 32 DNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLP-SLANLSSLTDVNLGSN 90
+ R+ ++LS KL GTLP L S L TL N L GP+P SL + SLT + +G N
Sbjct: 831 NGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGEN 1010
Query: 91 NFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIF 150
+ P GL L + L + N FP + + NL I ++ +++G LP
Sbjct: 1011 FLNGSIPRGLFGLPKLTQVELQD--NYLSGEFPEVGSVAVNLGQITLSNNQLSGVLPPSI 1184
Query: 151 GSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITVISNMTLLTQVWLH 210
G+FSS+ L L N +G +P I + L+++
Sbjct: 1185 GNFSSVQKLILDGNMFTGRIPPQ------------------------IGRLQQLSKIDFS 1292
Query: 211 VNKFTGPI-PDLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFG 269
NKF+GPI P++SQC + L L N+L+G +P+ + M L + L N L G +P
Sbjct: 1293 GNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSRNHLVGGIPSSI 1472
Query: 270 KDVK-YNSDDISHNN 283
++ S D S+NN
Sbjct: 1473 SSMQSLTSVDFSYNN 1517
Score = 84.7 bits (208), Expect = 2e-16
Identities = 77/283 (27%), Positives = 128/283 (45%), Gaps = 4/283 (1%)
Frame = +3
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAG-SGIQSFWINNNLPGLT--GSITVI 198
++G L L+ L LA N SG +P SL+ SG++ ++NN+ T ++ +
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRL 182
Query: 199 SNMTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLR 257
N+ +L L+ N TG +P ++Q +++ L L N +G +P LQ + +
Sbjct: 183 QNLEVLD---LYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVS 353
Query: 258 NNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSW 317
N+L+G +P P + L + G+ ++
Sbjct: 354 GNELEGTIP-------------------------PEIGNLSSLRELYIGY------YNTY 440
Query: 318 TGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
TG P + I ++ +L+ A GL G I A L L L+L N L+GS+
Sbjct: 441 TGGIPPE-----IGNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPE 605
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHG 420
L L L+++D+SNN LSGE+P ++K IT N++ K HG
Sbjct: 606 LGNLKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFRNKLHG 734
>TC233892 similar to UP|Q84P72 (Q84P72) Receptor-like protein kinase-like
protein (Fragment), partial (21%)
Length = 707
Score = 239 bits (610), Expect = 4e-63
Identities = 123/226 (54%), Positives = 155/226 (68%)
Frame = +2
Query: 666 QQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGR 725
+Q FIHRDLK SNILLGDD RAKV+DFGLVK APDG SV T+LAGTFGYLAPEYA TG+
Sbjct: 5 EQIFIHRDLKSSNILLGDDFRAKVSDFGLVKLAPDGKKSVVTRLAGTFGYLAPEYAVTGK 184
Query: 726 VTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPD 785
VTTK DV++FGVVLMEL+TG ALD+ P+E+ +L +WF + ++KE + AID LD
Sbjct: 185 VTTKADVFSFGVVLMELLTGLMALDEDRPEETQYLASWFWHIKSDKEKLMSAIDPALDIK 364
Query: 786 EETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQMS 845
EE + +AELAGHC+ R P QRPD+ HAVNVL PLVQ+W+P E D +
Sbjct: 365 EEMFDVVSIIAELAGHCSAREPNQRPDMSHAVNVLSPLVQKWKPLDDETEEYSGIDYSLP 544
Query: 846 LTQALQRWQANEGTSTFFNGMTSQTQSSSTSKPPVFADSLHSPDCR 891
L Q ++ WQ EG + + ++SS ++P FA+S S D R
Sbjct: 545 LNQMVKDWQETEGKDLSYVDL-QDSKSSIPARPTGFAESFTSVDGR 679
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 229 bits (585), Expect = 3e-60
Identities = 128/304 (42%), Positives = 181/304 (59%), Gaps = 3/304 (0%)
Frame = +1
Query: 545 SDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVA 604
S N++G GGFG+VY+G L DG K+A+K M G +G EF+ E+ +L+++ +L+A
Sbjct: 1 SKSNVIGHGGFGLVYRGVLNDGRKVAIKFMDQA--GKQGEEEFKVEVELLSRLHSPYLLA 174
Query: 605 LLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTP--LTWKQRLIIALDVGRGVEYLH 662
LLGYC + N +LLVYE M G L +HL+ TP L W+ RL IAL+ +G+EYLH
Sbjct: 175 LLGYCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLH 354
Query: 663 SLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GNYSVETKLAGTFGYLAPEYA 721
IHRD K SNILL AKV+DFGL K PD V T++ GT GY+APEYA
Sbjct: 355 EHVSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYA 534
Query: 722 ATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQT 781
TG +TTK DVY++GVVL+EL+TGR +D P LV+W +LT++E + K +D +
Sbjct: 535 LTGHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPS 714
Query: 782 LDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADD 841
L+ + +M + +VA +A C RP + V L PLV+ + S+ +
Sbjct: 715 LE-GQYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSLVPLVKTQRSPSKVGSSSSFNS 891
Query: 842 NQMS 845
++S
Sbjct: 892 PKLS 903
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 222 bits (566), Expect = 5e-58
Identities = 128/297 (43%), Positives = 178/297 (59%), Gaps = 6/297 (2%)
Frame = +2
Query: 533 SIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAK-----GSKGLNEF 587
++ LR+ TN FS N+LG GGFG VYKG + D + +K K G +G E+
Sbjct: 38 TLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLDLDGLQGHREW 217
Query: 588 QAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQR 647
AEI L ++RH HLV L+GYC RLL+YE+MP+G+L LF R + + W R
Sbjct: 218 LAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLF--RRYS-AAMPWSTR 388
Query: 648 LIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VE 706
+ IAL +G+ +LH A + I+RD K SNILL D AK++DFGL K+ P+G + V
Sbjct: 389 MKIALGAAKGLAFLHE-ADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDGPEGEDTHVT 565
Query: 707 TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRR 766
T++ GT GY APEY TG +TTK DVY++GVVL+EL+TGR+ +D S +E LV W R
Sbjct: 566 TRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGKSLVEWARP 745
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPL 823
+L +++ + ID+ L+ + M KVA LA C + P RP + + VL PL
Sbjct: 746 LLRDQKKVYNIIDRRLE-GQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVIKVLEPL 913
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 218 bits (554), Expect = 1e-56
Identities = 132/333 (39%), Positives = 195/333 (57%), Gaps = 12/333 (3%)
Frame = +2
Query: 493 DLASVSNGYGGASSELQSQSSGD---HSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNI 549
D++S S AS + S+S G+ S+L F S H LR T +F D++
Sbjct: 212 DISSNSRS-SSASIPVTSRSEGEILQSSNLKSF----------SYHELRAATRNFRPDSV 358
Query: 550 LGRGGFGIVYKGELPD----GTKIAVKRMISVAK----GSKGLNEFQAEIGVLTKVRHRH 601
LG GGFG V+KG + + TK + ++++V K G +G E+ AEI L +++H +
Sbjct: 359 LGEGGFGSVFKGWIDEHSLAATKPGIGKIVAVKKLNQDGLQGHREWLAEINYLGQLQHPN 538
Query: 602 LVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYL 661
LV L+GYC RLLVYE MP+G++ HLF R + P +W R+ IAL +G+ +L
Sbjct: 539 LVKLIGYCFEDEHRLLVYEFMPKGSMENHLFR-RGSYFQPFSWSLRMKIALGAAKGLAFL 715
Query: 662 HSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGYLAPEY 720
HS + I+RD K SNILL + AK++DFGL ++ P G+ S V T++ GT GY APEY
Sbjct: 716 HS-TEHKVIYRDFKTSNILLDTNYNAKLSDFGLARDGPTGDKSHVSTRVMGTRGYAAPEY 892
Query: 721 AATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQ 780
ATG +TTK DVY+FGVVL+E+I+GR+A+D + P +LV W + L+NK + + +D
Sbjct: 893 LATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHNLVEWAKPYLSNKRRVFRVMDP 1072
Query: 781 TLDPDEETMLSIYKVAELAGHCTTRSPYQRPDI 813
L+ + + A LA C + P RP++
Sbjct: 1073RLE-GQYSQNRAQAAAALAMQCFSVEPKCRPNM 1168
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 217 bits (552), Expect = 2e-56
Identities = 120/288 (41%), Positives = 178/288 (61%), Gaps = 4/288 (1%)
Frame = +3
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
L+ T FS N L GGFG V++G LPDG IAVK+ + ++G EF +E+ VL+
Sbjct: 21 LQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLAS--TQGDKEFCSEVEVLSC 194
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
+HR++V L+G+C++ RLLVYE++ G+L H++ +++ L W R IA+ R
Sbjct: 195 AQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQN---VLEWSARQKIAVGAAR 365
Query: 657 GVEYLHSLAQQS-FIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGY 715
G+ YLH + +HRD++P+NILL D A V DFGL + PDG+ VET++ GTFGY
Sbjct: 366 GLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVETRVIGTFGY 545
Query: 716 LAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIP 775
LAPEYA +G++T K DVY+FG+VL+EL+TGRKA+D + P L W R +L K+
Sbjct: 546 LAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLL-EKQATY 722
Query: 776 KAIDQTLD---PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
K ID +L D+E +Y++ + + C R P+ RP + + +L
Sbjct: 723 KLIDPSLRNCYVDQE----VYRMLKCSSLCIGRDPHLRPRMSQVLRML 854
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 217 bits (552), Expect = 2e-56
Identities = 123/301 (40%), Positives = 177/301 (57%), Gaps = 11/301 (3%)
Frame = +1
Query: 531 TISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPD----------GTKIAVKRMISVAKG 580
+ ++ L+ T +F D++LG GGFG V+KG + + G IAVKR+ G
Sbjct: 241 SFTLSELKTATRNFRPDSVLGEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQ--DG 414
Query: 581 SKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYT 640
G E+ AE+ L + H HLV L+G+C+ RLLVYE MP+G+ HLF R +
Sbjct: 415 IPGHREWLAEVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFR-RGSYFQ 591
Query: 641 PLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD 700
PL+W RL +ALD +G+ +LHS A+ I+RD K SN+LL AK++DFGL K+ P
Sbjct: 592 PLSWSLRLKVALDAAKGLAFLHS-AEAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPT 768
Query: 701 GNYS-VETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSH 759
G+ S V T++ GT+GY APEY ATG +T K DVY+FGVVL+E+++G++A+D + P +
Sbjct: 769 GDKSHVSTRVMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHN 948
Query: 760 LVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNV 819
LV W + + NK I + +D L T YK+A LA C + RP++ V
Sbjct: 949 LVEWAKPFMANKRKIFRVLDTRLQGQYSTD-DAYKLATLALRCLSIESKFRPNMDQVVTT 1125
Query: 820 L 820
L
Sbjct: 1126L 1128
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 209 bits (531), Expect = 6e-54
Identities = 121/293 (41%), Positives = 172/293 (58%), Gaps = 9/293 (3%)
Frame = +1
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVK----RMISVAK----GSKGLNEFQ 588
L+ T +F D++LG GGFG VYKG + + T A K +++V K G +G E+
Sbjct: 454 LKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGLQGHKEWL 633
Query: 589 AEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRL 648
E+ L ++ H++LV L+GYC +G RLLVYE M +G+L HLF G PL+W R+
Sbjct: 634 TEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFR---RGPQPLSWSVRM 804
Query: 649 IIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VET 707
+A+ RG+ +LH+ A+ I+RD K SNILL + AK++DFGL K P G+ + V T
Sbjct: 805 KVAIGAARGLSFLHN-AKSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGDRTHVST 981
Query: 708 KLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRV 767
++ GT GY APEY ATGR+T K DVY+FGVVL+EL++GR+A+D S +LV W +
Sbjct: 982 QVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLVEWAKPY 1161
Query: 768 LTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
L +K + + +D L + Y A LA C R RP I + L
Sbjct: 1162LGDKRRLFRIMDTKLG-GQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTL 1317
>TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (66%)
Length = 1737
Score = 206 bits (525), Expect = 3e-53
Identities = 118/283 (41%), Positives = 170/283 (59%), Gaps = 5/283 (1%)
Frame = +3
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGT-KIAVKRMISVAKGSKGLNEFQAEIGVLT 595
++ TN+F + +LG GGFG VYKGE+ GT K+A+KR +++ +G++EFQ EI +L+
Sbjct: 414 IKAATNNFDEALLLGVGGFGKVYKGEIDGGTTKVAIKRGNPLSE--QGVHEFQTEIEMLS 587
Query: 596 KVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVG 655
K+RHRHLV+L+GYC E +LVY++M GTL +HL++ ++ P WKQRL I +
Sbjct: 588 KLRHRHLVSLIGYCEENTEMILVYDYMAYGTLREHLYKTQK---PPRPWKQRLEICIGAA 758
Query: 656 RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GNYSVETKLAGTFG 714
RG+ YLH+ A+ + IHRD+K +NILL + AKV+DFGL K P N V T + G+FG
Sbjct: 759 RGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFG 938
Query: 715 YLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENI 774
YL PEY ++T K DVY+FGVVL E++ R AL+ ++ E L W
Sbjct: 939 YLDPEYFRRQQLTDKSDVYSFGVVLFEVLCARPALNPTLAKEQVSLAEWAAHCYQK---- 1106
Query: 775 PKAIDQTLDPDEETMLS---IYKVAELAGHCTTRSPYQRPDIG 814
+D +DP + ++ K AE A C RP +G
Sbjct: 1107-GILDSIIDPYLKGKIAPECFKKFAETAMKCVADQGIDRPSMG 1232
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 126 bits (316), Expect(2) = 7e-53
Identities = 72/168 (42%), Positives = 96/168 (56%), Gaps = 2/168 (1%)
Frame = +3
Query: 671 HRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGYLAPEYAATGRVTTK 729
+RD K SNILL + K++DFGL K P G+ S V T++ GT+GY APEYA TG++T K
Sbjct: 657 NRDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVK 836
Query: 730 VDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETM 789
DVY+FGVV +ELITGRKA+D + P +LVTW R + ++ K D L M
Sbjct: 837 SDVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRLQ-GRFPM 1013
Query: 790 LSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ-WEPTTHTDES 836
+Y+ +A C S RP IG V L L Q ++P + S
Sbjct: 1014RGLYQALAVASMCIQESAATRPLIGDVVTALSYLANQAYDPNGYRGSS 1157
Score = 100 bits (250), Expect(2) = 7e-53
Identities = 55/145 (37%), Positives = 81/145 (54%)
Frame = +2
Query: 531 TISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAE 590
T + L T +F ++ +G GGFG VYKG L +I + + G +G EF E
Sbjct: 242 TFTFRELAAATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLD-KNGLQGNREFLVE 418
Query: 591 IGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLII 650
+ +L+ + H +LV L+GYC +G++RLLVYE MP G+L HL + PL W R+ I
Sbjct: 419 VLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDL-PPDKEPLDWNTRMKI 595
Query: 651 ALDVGRGVEYLHSLAQQSFIHRDLK 675
A+ +G+EYLH A I + L+
Sbjct: 596 AVGAAKGLEYLHDKANPPGIQQRLQ 670
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 204 bits (520), Expect = 1e-52
Identities = 123/313 (39%), Positives = 175/313 (55%), Gaps = 9/313 (2%)
Frame = +1
Query: 510 SQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPD---- 565
S SSGD +G + L+ T +F D +LG GGFG V+KG L +
Sbjct: 46 SVSSGDQPYPNGQILPTSNLRIFTFAELKAATRNFKADTVLGEGGFGKVFKGWLEEKATS 225
Query: 566 ----GTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEH 621
GT IAVK++ S + +GL E+Q+E+ L ++ H +LV LLGYC+ +E LLVYE
Sbjct: 226 KGGSGTVIAVKKLNS--ESLQGLEEWQSEVNFLGRLSHTNLVKLLGYCLEESELLLVYEF 399
Query: 622 MPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILL 681
M +G+L HLF R PL W RL IA+ RG+ +LH+ + I+RD K SNILL
Sbjct: 400 MQKGSLENHLFG-RGSAVQPLPWDIRLKIAIGAARGLAFLHT--SEKVIYRDFKASNILL 570
Query: 682 GDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLM 740
AK++DFGL K P + S V T++ GT GY APEY ATG + K DVY FGVVL+
Sbjct: 571 DGSYNAKISDFGLAKLGPSASQSHVTTRVMGTHGYAAPEYVATGHLYVKSDVYGFGVVLV 750
Query: 741 ELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAG 800
E++TG +ALD + P L W + L ++ + +D L+ + + +++A+L+
Sbjct: 751 EILTGLRALDSNRPSGQHKLTEWVKPYLHDRRKLKGIMDSRLE-GKFPSKAAFRIAQLSM 927
Query: 801 HCTTRSPYQRPDI 813
C P RP +
Sbjct: 928 KCLASEPKHRPSM 966
>TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {Arabidopsis
thaliana;} , partial (42%)
Length = 1533
Score = 204 bits (518), Expect = 2e-52
Identities = 119/313 (38%), Positives = 178/313 (56%), Gaps = 8/313 (2%)
Frame = +2
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKG-LNEFQAEIGVLT 595
L T++F+ DN++GRGG VY+G LPDG ++AVK + K S+ + EF EI ++T
Sbjct: 437 LLSATSNFASDNLIGRGGCSYVYRGCLPDGKELAVK----ILKPSENVIKEFVQEIEIIT 604
Query: 596 KVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLF----ECREHGYTPLTWKQRLIIA 651
+RH++++++ G+C+ GN LLVY+ + +G+L ++L +C G W++R +A
Sbjct: 605 TLRHKNIISISGFCLEGNHLLLVYDFLSRGSLEENLHGNKVDCSAFG-----WQERYKVA 769
Query: 652 LDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAG 711
+ V ++YLH+ Q+ IHRD+K SNILL DD +++DFGL ++ T +AG
Sbjct: 770 VGVAEALDYLHNGCAQAVIHRDVKSSNILLADDFEPQLSDFGLASWGSSSSHITCTDVAG 949
Query: 712 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNK 771
TFGYLAPEY GRVT K+DVYAFGVVL+EL++ RK +++ P LV W +L
Sbjct: 950 TFGYLAPEYFMHGRVTDKIDVYAFGVVLLELLSNRKPINNESPKGQESLVMWATPILEGG 1129
Query: 772 ENIPKAIDQTLDP---DEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWE 828
+ Q LDP E I ++ A C R P RP I A L L
Sbjct: 1130K-----FSQLLDPSLGSEYNTCQIKRMILAATLCIRRIPRLRPQINLA---LLDLEDDTV 1285
Query: 829 PTTHTDESTCADD 841
+ T++S +D
Sbjct: 1286SISSTEQSVSLED 1324
>TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (8%)
Length = 1250
Score = 202 bits (513), Expect = 7e-52
Identities = 119/285 (41%), Positives = 181/285 (62%), Gaps = 1/285 (0%)
Frame = +1
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
L ++TN+F+ ILGRGGFG VY G + D T++AVK + A +G +F AE+ +L +
Sbjct: 64 LVKITNNFT--RILGRGGFGKVYHGFIDD-TQVAVKMLSPSAV--RGYEQFLAEVKLLMR 228
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
V HR+L +L+GYC N L+YE+M G L + + + LTW+ RL IALD +
Sbjct: 229 VHHRNLTSLVGYCNEENNIGLIYEYMANGNLDE-IVSGKSSRAKFLTWEDRLQIALDAAQ 405
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGNYSVETKLAGTFGY 715
G+EYLH+ + IHRD+K +NILL ++ +AK+ADFGL K+ P DG + T +AGT GY
Sbjct: 406 GLEYLHNGCKPPIIHRDVKCANILLNENFQAKLADFGLSKSFPTDGGSYMSTVVAGTPGY 585
Query: 716 LAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIP 775
L PEY+ + R+T K DVY+FGVVL+E++TG+ A+ PD+ +H+ W + +L+N + I
Sbjct: 586 LDPEYSISSRLTEKSDVYSFGVVLLEMVTGQPAI-AKTPDK-THISQWVKSMLSNGD-IK 756
Query: 776 KAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
D D +T S++++ E+ + SP++RP + + VN L
Sbjct: 757 NIADSRFKEDFDTS-SVWRIVEIGMASVSISPFKRPSMSYIVNEL 888
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 201 bits (512), Expect = 9e-52
Identities = 127/312 (40%), Positives = 177/312 (56%), Gaps = 1/312 (0%)
Frame = +1
Query: 555 FGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNE 614
FG VYKG L DGT +AVKR + +GL EF+ EI +L+K+RHRHLV+L+GYC +E
Sbjct: 1 FGRVYKGTLEDGTNVAVKR--GNPRSEQGLAEFRTEIEMLSKLRHRHLVSLIGYCDERSE 174
Query: 615 RLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDL 674
+LVYE+M G L HL+ PL+WKQRL I + RG+ YLH+ A QS IHRD+
Sbjct: 175 MILVYEYMANGPLRSHLYGT---DLPPLSWKQRLEICIGAARGLHYLHTGASQSIIHRDV 345
Query: 675 KPSNILLGDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGYLAPEYAATGRVTTKVDVY 733
K +NIL+ D+ AKVADFGL K P + + V T + G+FGYL PEY ++T K DVY
Sbjct: 346 KTTNILVDDNFVAKVADFGLSKTGPALDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVY 525
Query: 734 AFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIY 793
+FGVVLME++ R AL+ +P E ++ W K + + +DQ L + S+
Sbjct: 526 SFGVVLMEVLCTRPALNPVLPREQVNIAEW-AMSWQKKGMLDQIMDQNL-VGKVNPASLK 699
Query: 794 KVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRW 853
K E A C RP +G + L +Q E ++ E N ++ Q L R
Sbjct: 700 KFGETAEKCLAEYGVDRPSMGDVLWNLEYALQLQETSSALMEPEDNSTNHITGIQ-LTRL 876
Query: 854 QANEGTSTFFNG 865
+ + T + +G
Sbjct: 877 EPFDNTVSMIDG 912
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,998,734
Number of Sequences: 63676
Number of extensions: 580781
Number of successful extensions: 6538
Number of sequences better than 10.0: 1151
Number of HSP's better than 10.0 without gapping: 4775
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5492
length of query: 891
length of database: 12,639,632
effective HSP length: 106
effective length of query: 785
effective length of database: 5,889,976
effective search space: 4623631160
effective search space used: 4623631160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146585.14


