

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.12 + phase: 0 /pseudo
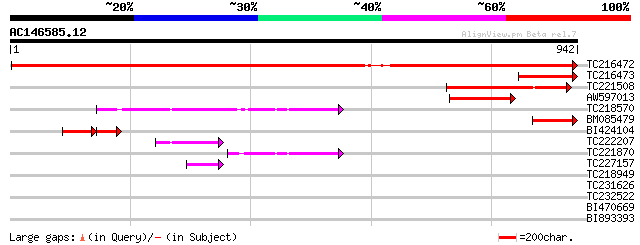
(942 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogena... 1548 0.0
TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserin... 187 2e-47
TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-... 172 8e-43
AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4)... 169 4e-42
TC218570 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctiona... 153 3e-37
BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2... 141 1e-33
BI424104 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2... 97 3e-33
TC222207 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctiona... 67 4e-11
TC221870 similar to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional ... 60 6e-09
TC227157 similar to UP|Q40329 (Q40329) Serine proteinase inhibit... 50 4e-06
TC218949 weakly similar to UP|Q7V6C2 (Q7V6C2) Uridylate kinase ... 42 0.002
TC231626 similar to UP|Q9VXX2 (Q9VXX2) CG9101-PA, partial (10%) 33 0.59
TC232522 similar to UP|Q05680 (Q05680) Auxin-responsive GH3 prod... 31 2.2
BI470669 weakly similar to GP|9758160|dbj| contains similarity t... 31 2.2
BI893393 similar to GP|9800656|gb| 1-deoxy-D-xylulose-5-phosphat... 30 3.8
>TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogenase ,
complete
Length = 3148
Score = 1548 bits (4009), Expect = 0.0
Identities = 794/941 (84%), Positives = 852/941 (90%), Gaps = 1/941 (0%)
Frame = +2
Query: 3 SSLSSSLSHFSRISVTSLQ-HDYNNKIPADSQCRHFLLSRRFHSLRKGITLPRRRESPSS 61
+S S++++ FSR+S T H +++ SQCR F LSR HSLRKG+TLPR RE+PS+
Sbjct: 32 ASFSAAVAQFSRVSPTHTPLHSHSHDTLFQSQCRPFFLSRTSHSLRKGLTLPRGREAPST 211
Query: 62 GICASLTDVSVNVAVEEKELSKGDSWSVHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVV 121
+ AS TDVS NV++EEK+L KG++WSVHKFGGTC+G+SQRIKNV DI+L DDSERKLVV
Sbjct: 212 TVRASFTDVSPNVSLEEKQLPKGETWSVHKFGGTCVGTSQRIKNVADIILKDDSERKLVV 391
Query: 122 VSAMSKVTDMMYDLINKAQSRDESYISSLDAVLEKHSATAHDILDGETLAIFLSKLHEDI 181
VSAMSKVTDMMYDLI+KAQSRDESY ++L+AV EKHSATAHDILDG+ LA FLSKLH DI
Sbjct: 392 VSAMSKVTDMMYDLIHKAQSRDESYTAALNAVSEKHSATAHDILDGDNLATFLSKLHHDI 571
Query: 182 SNLKAMLRAIYIAGHVTESFTDFVVGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVNP 241
SNLKAMLRAIYIAGH TESFTDFVVGHGELWSAQMLSLVI KNG DCKWMDTR+VLIVNP
Sbjct: 572 SNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVITKNGTDCKWMDTRDVLIVNP 751
Query: 242 TSSNQVDPDYLESERRLEKWYSLNPCKVIIATGFIASTPENIPTTLKRDGSDFSAAIMGS 301
T SNQVDPDYLESE+RLEKWYSLNPCKVIIATGFIASTP+NIPTTLKRDGSDFSAAIMG+
Sbjct: 752 TGSNQVDPDYLESEQRLEKWYSLNPCKVIIATGFIASTPQNIPTTLKRDGSDFSAAIMGA 931
Query: 302 LFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRY 361
LF+ARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRY
Sbjct: 932 LFKARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRY 1111
Query: 362 GIPILIRNIFNLSAPGTKICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVP 421
GIPI+IRNIFNLSAPGTKICHP V+D+ED NLQN+VKGFATIDNLALVNVEGTGMAGVP
Sbjct: 1112GIPIMIRNIFNLSAPGTKICHPSVNDHEDSQNLQNFVKGFATIDNLALVNVEGTGMAGVP 1291
Query: 422 GTASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGRLSQ 481
GTASAIF AVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGRLSQ
Sbjct: 1292GTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGRLSQ 1471
Query: 482 VAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSI 541
VAVIPNCSILAAVGQKMASTPGVSA+LFNALAKANINVRAIAQGCSEYNITVV+KRED I
Sbjct: 1472VAVIPNCSILAAVGQKMASTPGVSASLFNALAKANINVRAIAQGCSEYNITVVVKREDCI 1651
Query: 542 KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQAKHSHLYISYLLHTRRLLEICG 601
KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQA + L R++ I G
Sbjct: 1652KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASTLKEEFNIDL---RVMGILG 1822
Query: 602 KLMQQFIQCRLQF*KKNLTLICV*WA*LAQSQCFSVMSKWKELREERGEVANLEKFAQHV 661
K++ L S +++W+ELREERGEVAN+EKF QHV
Sbjct: 1823--------------SKSMLL----------SDVGIDLARWRELREERGEVANMEKFVQHV 1930
Query: 662 HGNNFIPNTALVDCTADSIIAGHYYEWLCKGIHVITPNKKANSGPLEQYLRLRALQRQSY 721
HGN+FIPNTALVDCTADS IAG+YY+WL KGIHV+TPNKKANSGPL+QYL+LRALQRQSY
Sbjct: 1931HGNHFIPNTALVDCTADSAIAGYYYDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSY 2110
Query: 722 THYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVGEA 781
THYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVV EA
Sbjct: 2111THYFYEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVSEA 2290
Query: 782 KEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPLRACASAQEFMQ 841
KEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIP+ESLVPEPLRACASAQEFMQ
Sbjct: 2291KEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLRACASAQEFMQ 2470
Query: 842 QLPKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIA 901
+LPKFDQEF KKQEDA+NAGEVLRYVGVVDVTNKKGVVELR+YKKDHPFAQLSGSDNIIA
Sbjct: 2471ELPKFDQEFTKKQEDAENAGEVLRYVGVVDVTNKKGVVELRRYKKDHPFAQLSGSDNIIA 2650
Query: 902 FTTRRYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 942
FTTRRYK+QPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP
Sbjct: 2651FTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 2773
>TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserine
dehydrogenase , partial (11%)
Length = 604
Score = 187 bits (475), Expect = 2e-47
Identities = 92/97 (94%), Positives = 96/97 (98%)
Frame = +2
Query: 846 FDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIAFTTR 905
FDQEF KKQEDA+NAGEVLRYVGVVDVTN+KGVVELR+YKKDHPFAQLSGSDNIIAFTTR
Sbjct: 2 FDQEFTKKQEDAENAGEVLRYVGVVDVTNEKGVVELRRYKKDHPFAQLSGSDNIIAFTTR 181
Query: 906 RYKNQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 942
RYK+QPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP
Sbjct: 182 RYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAP 292
>TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-like
protein, partial (60%)
Length = 912
Score = 172 bits (435), Expect = 8e-43
Identities = 90/210 (42%), Positives = 136/210 (63%), Gaps = 2/210 (0%)
Frame = +2
Query: 726 YEATVGAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVGEAKEAG 785
+E+TVGAGLP++++L ++ +GD + QI G SGTL Y+ + +DG+ S+VV +AK G
Sbjct: 50 HESTVGAGLPVIASLNRIISSGDPVHQIIGSLSGTLGYVMSEVEDGKPLSQVVRDAKSLG 229
Query: 786 YTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPL-RACASAQEFMQQ-L 843
YTEPDPRDDL G DVARK +ILAR G ++ L +I IESL P+ + + ++F+ + L
Sbjct: 230 YTEPDPRDDLGGMDVARKALILARILGHQINLDSIQIESLYPKEMGPGVMTVEDFLNRGL 409
Query: 844 PKFDQEFAKKQEDADNAGEVLRYVGVVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIAFT 903
D++ + E A + G VLRYV V + + V +++ K+ P +L GSDN++
Sbjct: 410 LLLDKDIQDRVEKAASNGNVLRYVCV--IMGSRCEVGIQELPKNSPLGRLRGSDNVLEIY 583
Query: 904 TRRYKNQPLIVRGPGAGAQVTAGGIFSDIL 933
TR Y QPL+++G GAG TA G+ +DI+
Sbjct: 584 TRCYSKQPLVIQGAGAGNDTTAAGVLADIV 673
>AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (12%)
Length = 334
Score = 169 bits (429), Expect = 4e-42
Identities = 87/110 (79%), Positives = 95/110 (86%)
Frame = +2
Query: 731 GAGLPIVSTLRGLLETGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVGEAKEAGYTEPD 790
G GLPIVSTLRGLLETGDKILQIEGIF GTLSYIFNNFKDGRA+SE E+KEA YT+P+
Sbjct: 5 GPGLPIVSTLRGLLETGDKILQIEGIFIGTLSYIFNNFKDGRAYSEESSESKEARYTDPN 184
Query: 791 PRDDLSGTDVARKVIILARESGLKLELSNIPIESLVPEPLRACASAQEFM 840
P DDLSGTD A K IILA+ESGLKLEL+NIP +SL PEPL ACA + EFM
Sbjct: 185 PIDDLSGTDAAIKAIILAKESGLKLELTNIPDDSLEPEPLLACA*SHEFM 334
>TC218570 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (75%)
Length = 1493
Score = 153 bits (387), Expect = 3e-37
Identities = 109/416 (26%), Positives = 212/416 (50%), Gaps = 6/416 (1%)
Frame = +1
Query: 145 SYISSLDAVLEKHSATAHDI-LDGETLAIFLSKLHEDISNLKAMLRAIYIAGHVTESFTD 203
S I L + + H T + +DG +A L +L + +L+ I + +T+ D
Sbjct: 19 SSIEELCFIKDLHLRTVDQLGVDGSVIAKHLEELEQ-------LLKGIAMMKELTKRTQD 177
Query: 204 FVVGHGELWSAQMLSLVIRKNGIDCKWMDTREVLIVNP---TSSNQVDPDYLESERRLEK 260
++V GE S ++ + + K G+ + D E+ + T+++ ++ Y +RL
Sbjct: 178 YLVSFGECMSTRIFAAYLNKIGVKARQYDAFEIGFITTDDFTNADILEATYPAVAKRLHG 357
Query: 261 WYSLNPCKVIIATGFIASTPENIP-TTLKRDGSDFSAAIMGSLFRARQVTIWTDVDGVYS 319
+ +P + I TGF+ ++ TTL R GSD +A +G ++ +W DVDGV +
Sbjct: 358 DWLSDPA-IAIVTGFLGKARKSCAVTTLGRGGSDLTATTIGKALGLPEIQVWKDVDGVLT 534
Query: 320 ADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMRYGIPILIRNIFNLSAPGTK 379
DP +A + L++ EA E++YFGA VLHP+++ P IP+ ++N +N APGT
Sbjct: 535 CDPNIYPKAEPVPYLTFDEAAELAYFGAQVLHPQSMRPARESDIPVRVKNSYNPKAPGT- 711
Query: 380 ICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPGTASAIFAAVKDVGANVI 439
+++ D S + + N+ ++++ T M G G + +F+ +++G +V
Sbjct: 712 ----LITKARDMS--KAVLTSIVLKRNVTMLDIASTRMLGQYGFLAKVFSIFEELGISVD 873
Query: 440 MISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDN-GRLSQVAVIPNCSILAAVGQKM 498
++ A+SE SV + ++ + E +Q ++ +++ V ++ N SI++ +G
Sbjct: 874 VV--ATSEVSVSLTLDPSKLWS-RELIQQELDHVVEELEKIAVVNLLQNRSIISLIGNVQ 1044
Query: 499 ASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSIKALRAVHSRFYLS 554
S+ + F L + V+ I+QG S+ NI++V+ ++ + +RA+HS F+ S
Sbjct: 1045RSSL-ILEKAFRVLRTLGVTVQMISQGASKVNISLVVNDSEAEQCVRALHSAFFES 1209
>BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (8%)
Length = 347
Score = 141 bits (356), Expect = 1e-33
Identities = 68/74 (91%), Positives = 74/74 (99%)
Frame = +1
Query: 869 VVDVTNKKGVVELRKYKKDHPFAQLSGSDNIIAFTTRRYKNQPLIVRGPGAGAQVTAGGI 928
VVDV+N+KGVVELR+YKKDHPFAQLSGSDNIIAFTTRRY+NQPLIVRGPGAGAQVTAGGI
Sbjct: 1 VVDVSNEKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYRNQPLIVRGPGAGAQVTAGGI 180
Query: 929 FSDILRLASYLGAP 942
F+DILRLA+YLGAP
Sbjct: 181 FNDILRLATYLGAP 222
>BI424104 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (10%)
Length = 421
Score = 96.7 bits (239), Expect(2) = 3e-33
Identities = 47/56 (83%), Positives = 52/56 (91%)
Frame = +1
Query: 89 VHKFGGTCMGSSQRIKNVGDIVLNDDSERKLVVVSAMSKVTDMMYDLINKAQSRDE 144
VHKFGGTC+G+SQRIKNV DI+L DDS RKLVVVSAMSKV +MMYDLI+KAQSRDE
Sbjct: 1 VHKFGGTCVGTSQRIKNVADIILKDDSGRKLVVVSAMSKVKNMMYDLIHKAQSRDE 168
Score = 65.1 bits (157), Expect(2) = 3e-33
Identities = 31/41 (75%), Positives = 36/41 (87%)
Frame = +3
Query: 145 SYISSLDAVLEKHSATAHDILDGETLAIFLSKLHEDISNLK 185
+YI++LD+V EKH+ATAHDILDG LA FLSKLH DISNLK
Sbjct: 168 AYIAALDSV*EKHNATAHDILDGHNLASFLSKLHHDISNLK 290
>TC222207 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (20%)
Length = 360
Score = 67.0 bits (162), Expect = 4e-11
Identities = 38/114 (33%), Positives = 64/114 (55%), Gaps = 1/114 (0%)
Frame = +3
Query: 242 TSSNQVDPDYLESERRLEKWYSLNPCKVIIATGFIASTPENIP-TTLKRDGSDFSAAIMG 300
T+++ ++ Y +RL + +P + I TGF+ ++ TTL R SD +A +G
Sbjct: 21 TNADILEATYPAVAKRLHSDWVSDPA-IPIVTGFLGKARKSCAVTTLGRG*SDLTATTIG 197
Query: 301 SLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRT 354
++ +W DVDGV + DP +A + L++ EA E++YFGA VLHP++
Sbjct: 198 KALGLPEIQVWKDVDGVLTCDPNICPQAKPVPYLTFDEAAELAYFGAXVLHPQS 359
>TC221870 similar to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (37%)
Length = 882
Score = 59.7 bits (143), Expect = 6e-09
Identities = 43/193 (22%), Positives = 97/193 (49%), Gaps = 1/193 (0%)
Frame = +2
Query: 363 IPILIRNIFNLSAPGTKICHPVVSDYEDKSNLQNYVKGFATIDNLALVNVEGTGMAGVPG 422
IP+ ++N N APGT I + + + + N+ ++++ T M G G
Sbjct: 14 IPVRVKNXXNXKAPGTLIA-------KTRDMSKALLTSIVLKRNVTMLDIVSTRMLGQFG 172
Query: 423 TASAIFAAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDN-GRLSQ 481
+ +F+ +++G +V ++ A+SE S+ + ++ + E +Q ++ +++
Sbjct: 173 FLAKVFSIFEELGISVDVV--ATSEVSISLTLDPSKLWS-RELIQQELDYVVEELEKIAV 343
Query: 482 VAVIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVIKREDSI 541
V ++ SI++ +G S+ + F+ L + V+ I+QG S+ NI++V+ ++
Sbjct: 344 VNLLKTRSIISLIGNVQRSSL-ILEKAFHVLRTLGVTVQMISQGASKVNISLVVNDSEAE 520
Query: 542 KALRAVHSRFYLS 554
+ +RA+H F+ S
Sbjct: 521 QCVRALHKAFFES 559
>TC227157 similar to UP|Q40329 (Q40329) Serine proteinase inhibitor, partial
(89%)
Length = 769
Score = 50.4 bits (119), Expect = 4e-06
Identities = 25/61 (40%), Positives = 36/61 (58%)
Frame = -3
Query: 294 FSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPR 353
+ ++GS A V I TD+ GV +ADPR V AV L L++ EA E++ +G + H R
Sbjct: 185 YKPTLVGSSLGADYVLINTDISGVMTADPRIVDRAVALSHLNHHEALELAVYGTRMFHAR 6
Query: 354 T 354
T
Sbjct: 5 T 3
>TC218949 weakly similar to UP|Q7V6C2 (Q7V6C2) Uridylate kinase , partial
(92%)
Length = 1167
Score = 41.6 bits (96), Expect = 0.002
Identities = 27/86 (31%), Positives = 45/86 (51%)
Frame = +1
Query: 292 SDFSAAIMGSLFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLH 351
+D +AA+ + A V T+VDGV+ DP++ +A + +TL+YQE +V+
Sbjct: 547 TDTAAALRCAEMNAEVVLKATNVDGVFDEDPKRNPQARLHETLTYQEVTSKD---LSVMD 717
Query: 352 PRTIIPVMRYGIPILIRNIFNLSAPG 377
I IP++ +FNL+ PG
Sbjct: 718 MTAITLCQENNIPVI---VFNLNKPG 786
>TC231626 similar to UP|Q9VXX2 (Q9VXX2) CG9101-PA, partial (10%)
Length = 663
Score = 33.1 bits (74), Expect = 0.59
Identities = 20/80 (25%), Positives = 35/80 (43%)
Frame = +3
Query: 212 WSAQMLSLVIRKNGIDCKWMDTREVLIVNPTSSNQVDPDYLESERRLEKWYSLNPCKVII 271
W + + V KNG + +W++ R+ + N +++ D + RR KW
Sbjct: 435 WFKECYATVTEKNGDEYEWVENRD*RLENAMRKIKIEKDIIIENRR--KWRERE------ 590
Query: 272 ATGFIASTPENIPTTLKRDG 291
G S+ + + T L RDG
Sbjct: 591 CNGIA*SSTQQLETPLMRDG 650
>TC232522 similar to UP|Q05680 (Q05680) Auxin-responsive GH3 product, partial
(55%)
Length = 1006
Score = 31.2 bits (69), Expect = 2.2
Identities = 17/35 (48%), Positives = 20/35 (56%)
Frame = +2
Query: 29 PADSQCRHFLLSRRFHSLRKGITLPRRRESPSSGI 63
P S R L + R H LR + LPRRR SPS G+
Sbjct: 692 PCRS*TRKILRAHRHHVLRS-VPLPRRRHSPSHGV 793
>BI470669 weakly similar to GP|9758160|dbj| contains similarity to
retroelement pol polyprotein~gene_id:MOK9.20
{Arabidopsis thaliana}, partial (3%)
Length = 415
Score = 31.2 bits (69), Expect = 2.2
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = -3
Query: 581 KHSHLYISYLLHTRRLLEICGKLMQQFIQC 610
KH H+++ + LH+ L IC L+ FI C
Sbjct: 110 KHGHMFLLFFLHSLFRLHICTSLIVPFILC 21
>BI893393 similar to GP|9800656|gb| 1-deoxy-D-xylulose-5-phosphate
reductoisomerase {Artemisia annua}, partial (30%)
Length = 430
Score = 30.4 bits (67), Expect = 3.8
Identities = 19/54 (35%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Frame = -3
Query: 146 YISSLDAVLEKHSATAHDILD--GETLAIFLSKLHEDISNLKA-MLRAIYIAGH 196
YI + KH T+ +L E++ IFL KLH N A ++ I+ AGH
Sbjct: 383 YIQITNLFFNKHLITSRPLLKRVAESIVIFLPKLHMGCFNASASLISFIFSAGH 222
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,245,295
Number of Sequences: 63676
Number of extensions: 500000
Number of successful extensions: 2160
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 2152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2159
length of query: 942
length of database: 12,639,632
effective HSP length: 106
effective length of query: 836
effective length of database: 5,889,976
effective search space: 4924019936
effective search space used: 4924019936
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146585.12


