

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
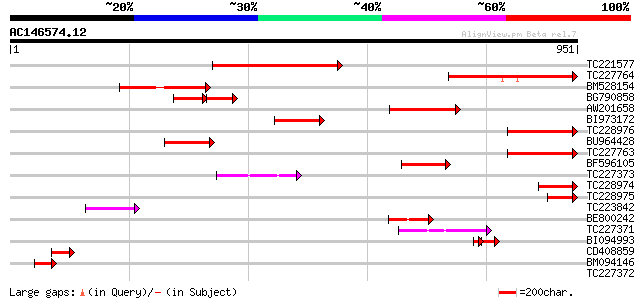
Query= AC146574.12 - phase: 0 /pseudo
(951 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221577 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, part... 316 3e-86
TC227764 243 3e-64
BM528154 230 3e-60
BG790858 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 101 1e-41
AW201658 157 3e-38
BI973172 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabido... 138 1e-32
TC228976 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, part... 136 5e-32
BU964428 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 133 3e-31
TC227763 125 1e-28
BF596105 107 2e-23
TC227373 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g2... 97 4e-20
TC228974 91 2e-18
TC228975 85 1e-16
TC223842 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g2... 72 2e-12
BE800242 67 4e-11
TC227371 similar to GB|AAQ89612.1|37201994|BT010590 At5g26850 {A... 65 1e-10
BI094993 52 8e-10
CD408859 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {... 56 9e-08
BM094146 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabido... 54 4e-07
TC227372 similar to GB|AAQ89612.1|37201994|BT010590 At5g26850 {A... 32 1.0
>TC221577 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, partial (13%)
Length = 775
Score = 316 bits (810), Expect = 3e-86
Identities = 161/218 (73%), Positives = 185/218 (84%), Gaps = 1/218 (0%)
Frame = +3
Query: 341 LVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEA 400
LVKHLDHKNVAKQPILQI+IIN TT++AQN+KQQASVA++GAISDLIKHLR+CLQNSAEA
Sbjct: 3 LVKHLDHKNVAKQPILQINIINTTTKLAQNLKQQASVAILGAISDLIKHLRKCLQNSAEA 182
Query: 401 TDIGNDAHTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAV 460
+ GND LNT+LQ ++EMCIL LS KVGD GPI DLMAVVLEN+SS+ I+A TTISAV
Sbjct: 183 SSTGNDGLKLNTELQFALEMCILHLSKKVGDVGPILDLMAVVLENISSTAIIAGTTISAV 362
Query: 461 YQTAKLITSVPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWL 520
YQTAKLI S+PNV YH KAFPDALFHQLLLAMAHPD ET++GAHSI S+VLMPS SP L
Sbjct: 363 YQTAKLIMSIPNVSYHKKAFPDALFHQLLLAMAHPDHETRVGAHSIFSLVLMPSPFSPQL 542
Query: 521 DQK-KISKKVESDGLSIQHESLSGEDPLNGKPVEEKPI 557
DQK IS+KV S+ SIQHES G + +NGK +E K +
Sbjct: 543 DQKTNISQKVPSESFSIQHESFLGAEQINGKSMEGKAV 656
>TC227764
Length = 1110
Score = 243 bits (620), Expect = 3e-64
Identities = 135/254 (53%), Positives = 172/254 (67%), Gaps = 38/254 (14%)
Frame = +3
Query: 736 VVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPD 795
+++GS ED+V+AMK++SAV+LDD+QLKETVIS F+TKFSKL EDELSSIK QL+QGFSPD
Sbjct: 9 LIYGSQEDDVSAMKTMSAVKLDDKQLKETVISCFLTKFSKLSEDELSSIKKQLVQGFSPD 188
Query: 796 DAYPSGPPLFMETPRPGSPLAQIEFPDVDE--------------------------MKGV 829
DAYP GPPLFMETP SPLAQIEFPD DE +
Sbjct: 189 DAYPLGPPLFMETPGKSSPLAQIEFPDFDEIVAPLALMDEETQPEPSGSQSDRKSSLSSN 368
Query: 830 GPN*VEASQIVEHHFQPTAQM-----------F*VLINSWNRLETGKQQKMLTIRSFKNQ 878
P+ + +Q+++ + Q+ + + N L TGKQQKM + SFK+Q
Sbjct: 369 SPDILSVNQLLQSVLETARQVASFPISSTPVPYDQMKNQCEALVTGKQQKMSILHSFKHQ 548
Query: 879 QETKAIVLSSENE-EVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRL 937
QET+A+VLSSENE +VS P+K L+YS+GDLKLV+Q+ QAQ Q+R S D +QHSL+L
Sbjct: 549 QETRALVLSSENETKVSPLPIKTLDYSEGDLKLVSQQPIQAQYQVRLCSYDFGQQHSLKL 728
Query: 938 PPSSPYDKFLKAAG 951
PP+SP+DKFLKAAG
Sbjct: 729 PPASPFDKFLKAAG 770
>BM528154
Length = 428
Score = 230 bits (586), Expect = 3e-60
Identities = 120/154 (77%), Positives = 129/154 (82%)
Frame = +2
Query: 184 LQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFP 243
LQ LS MV+FMGEHSHLSMDFDKIIS ILEN+ DLQSKSNLAKVEKLNSQSQ+QLVQ FP
Sbjct: 2 LQALSHMVQFMGEHSHLSMDFDKIISVILENFKDLQSKSNLAKVEKLNSQSQSQLVQGFP 181
Query: 244 KEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYF 303
KE G ESKLD AK+PAYWSK+CLYNIAKLAKEATTVRRVL+PLFH F
Sbjct: 182 KE------------GAVTESKLDAAKDPAYWSKLCLYNIAKLAKEATTVRRVLKPLFHNF 325
Query: 304 DTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLM 337
D+EN WSSEKGVA CVLMYLQ LLAESG+NSHL+
Sbjct: 326 DSENQWSSEKGVASCVLMYLQSLLAESGDNSHLL 427
>BG790858 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (11%)
Length = 417
Score = 101 bits (252), Expect(2) = 1e-41
Identities = 51/58 (87%), Positives = 53/58 (90%)
Frame = +3
Query: 275 SKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGN 332
SKVCL NIAKLAKEATTVRRVLE LFH FD+ENHWSSEKGVA CVLMYLQ LLAESG+
Sbjct: 3 SKVCLSNIAKLAKEATTVRRVLELLFHNFDSENHWSSEKGVASCVLMYLQSLLAESGS 176
Score = 88.2 bits (217), Expect(2) = 1e-41
Identities = 43/51 (84%), Positives = 48/51 (93%)
Frame = +2
Query: 331 GNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVAVIG 381
G+NSHL+LS LVKHLDHKNVAK+PILQIDIIN T Q+AQNVKQQASVA+IG
Sbjct: 263 GDNSHLLLSSLVKHLDHKNVAKKPILQIDIINTTMQLAQNVKQQASVAIIG 415
>AW201658
Length = 398
Score = 157 bits (396), Expect = 3e-38
Identities = 88/120 (73%), Positives = 98/120 (81%), Gaps = 1/120 (0%)
Frame = +3
Query: 638 IALLYYLLVLS-YMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAA 696
IALL+ S YMAL RCFQLAFSLRSISLDQEGGL PS RRSL TLAS+MLIFSARA
Sbjct: 39 IALLFSRSKASNYMALARCFQLAFSLRSISLDQEGGLQPSHRRSLFTLASYMLIFSARAG 218
Query: 697 DFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQL 756
+ LIP+VKASLTE VDPFLELVDD L+AVCI+S+K+ +GS EDEVAA KSLS V+L
Sbjct: 219 NVPGLIPEVKASLTEPTVDPFLELVDDIRLQAVCIESEKINYGSQEDEVAAAKSLSDVEL 398
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/21 (90%), Positives = 19/21 (90%)
Frame = +1
Query: 627 ALQIMRQWHTLIALLYYLLVL 647
ALQIMRQWHT IALLYY LVL
Sbjct: 1 ALQIMRQWHTPIALLYYFLVL 63
>BI973172 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (5%)
Length = 420
Score = 138 bits (347), Expect = 1e-32
Identities = 67/86 (77%), Positives = 78/86 (89%), Gaps = 1/86 (1%)
Frame = +2
Query: 444 ENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGA 503
EN+ +TI+AR+TI+AVYQTAKLITS+PNV YHNKAFPDALFHQLLLAMAHPD ETQIGA
Sbjct: 2 ENIPITTIIARSTITAVYQTAKLITSIPNVSYHNKAFPDALFHQLLLAMAHPDCETQIGA 181
Query: 504 HSILSMVLMPSVVSPWLDQK-KISKK 528
HS+ SMVLMPS+ SPWLD K KI+++
Sbjct: 182 HSVFSMVLMPSMFSPWLDHKTKIAQR 259
Score = 33.5 bits (75), Expect = 0.46
Identities = 19/44 (43%), Positives = 25/44 (56%)
Frame = +1
Query: 514 SVVSPWLDQKKISKKVESDGLSIQHESLSGEDPLNGKPVEEKPI 557
S+V P Q K + ++D S QHE+ SG + LNGK E K I
Sbjct: 223 SLVRP---QNKDCSEAQNDSFSTQHETFSGAENLNGKLEEGKAI 345
>TC228976 similar to UP|Q6YXW5 (Q6YXW5) Cyclin-like protein, partial (4%)
Length = 647
Score = 136 bits (342), Expect = 5e-32
Identities = 74/117 (63%), Positives = 84/117 (71%), Gaps = 1/117 (0%)
Frame = +1
Query: 836 ASQIVEHHFQPTAQMF*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENE-EVS 894
A Q+ T + + N L TGKQQKM I SFK+QQE+KAI+LSSENE +VS
Sbjct: 58 ARQVASFSTSSTPLPYDQMKNQCEALVTGKQQKMSVIHSFKHQQESKAIILSSENEVKVS 237
Query: 895 RQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
P KALEYS GDLKLVTQ+QF+ QDQ R RS D+ QHSLRLPPSSPYDKFLKAAG
Sbjct: 238 PLPAKALEYSNGDLKLVTQQQFEVQDQARHRSHDSGHQHSLRLPPSSPYDKFLKAAG 408
>BU964428 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (10%)
Length = 253
Score = 133 bits (335), Expect = 3e-31
Identities = 66/84 (78%), Positives = 75/84 (88%)
Frame = +2
Query: 260 EIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYCV 319
EI+ L+TAK+P YWSKVCLY++ KLA+EATT+RRVLEPLFHYFDTEN WSSEKGVA V
Sbjct: 2 EIDYVLNTAKDPTYWSKVCLYHMVKLAREATTLRRVLEPLFHYFDTENQWSSEKGVADHV 181
Query: 320 LMYLQFLLAESGNNSHLMLSILVK 343
LMYLQ LLAESG+NS L+LSILVK
Sbjct: 182 LMYLQSLLAESGDNSCLLLSILVK 253
>TC227763
Length = 717
Score = 125 bits (313), Expect = 1e-28
Identities = 69/117 (58%), Positives = 82/117 (69%), Gaps = 1/117 (0%)
Frame = +3
Query: 836 ASQIVEHHFQPTAQMF*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENE-EVS 894
A Q+ T + + N L TGKQQKM + SFK+QQET AIVLSSENE +VS
Sbjct: 36 ARQVASFPISSTPVSYDQMKNQCEALVTGKQQKMSILHSFKHQQETGAIVLSSENEIKVS 215
Query: 895 RQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
P+K LEYS+GDLKLV EQFQAQ Q+R S D +QHSL+LPP+SP+DKFLKAAG
Sbjct: 216 PLPIKTLEYSEGDLKLVHHEQFQAQYQVRLCSYDFGQQHSLKLPPASPFDKFLKAAG 386
>BF596105
Length = 246
Score = 107 bits (267), Expect = 2e-23
Identities = 55/82 (67%), Positives = 67/82 (81%)
Frame = +1
Query: 657 QLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKASLTEAPVDP 716
+LAFSL S+SLDQEGGL PSRRRSL TLAS+MLIFSARA +F +LI KVK SLTE VDP
Sbjct: 1 ELAFSLMSLSLDQEGGLQPSRRRSLFTLASYMLIFSARAGNFPELIQKVKTSLTETTVDP 180
Query: 717 FLELVDDNLLRAVCIKSDKVVF 738
LEL+DD L+AV +S+ +++
Sbjct: 181 XLELIDDVRLQAVSRESENIIY 246
>TC227373 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g26850
{Arabidopsis thaliana;} , partial (15%)
Length = 896
Score = 96.7 bits (239), Expect = 4e-20
Identities = 54/143 (37%), Positives = 86/143 (59%)
Frame = +1
Query: 347 HKNVAKQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGND 406
HKNV P L+ ++ + T +A ++ + +A I + L +HLR+ LQ S+E +G
Sbjct: 1 HKNVMNDPQLKTCVVQVATSLAMQIRSGSGLAEIVFVGVLCRHLRKSLQASSEF--VGEQ 174
Query: 407 AHTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKL 466
LN LQ+SI+ C+ +++N V DA P+FDLMA+ LEN+ S +V R TI ++ A+
Sbjct: 175 ELNLNISLQNSIDDCLQEIANGVIDAQPLFDLMAINLENIPSG-VVGRATIGSLIILARA 351
Query: 467 ITSVPNVLYHNKAFPDALFHQLL 489
+T + L+ + FP+AL QLL
Sbjct: 352 LTLALSHLHSQQGFPEALLVQLL 420
>TC228974
Length = 628
Score = 90.9 bits (224), Expect = 2e-18
Identities = 48/65 (73%), Positives = 52/65 (79%), Gaps = 1/65 (1%)
Frame = +3
Query: 888 SENE-EVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKF 946
SENE +VS KALEYS GDLKLVTQ+Q + QDQ R RS D+ QHSLRLPPSSPYDKF
Sbjct: 3 SENEVKVSPLXXKALEYSNGDLKLVTQQQXEVQDQARHRSHDSGHQHSLRLPPSSPYDKF 182
Query: 947 LKAAG 951
LKAAG
Sbjct: 183 LKAAG 197
>TC228975
Length = 387
Score = 85.1 bits (209), Expect = 1e-16
Identities = 40/50 (80%), Positives = 45/50 (90%)
Frame = +2
Query: 902 EYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
EYS GDLKLVTQ+QFQAQDQ R +S ++ +QHSLRLPPSSPYDKFLKAAG
Sbjct: 2 EYSNGDLKLVTQQQFQAQDQARHQSHESGQQHSLRLPPSSPYDKFLKAAG 151
>TC223842 weakly similar to GB|AAQ89612.1|37201994|BT010590 At5g26850
{Arabidopsis thaliana;} , partial (8%)
Length = 911
Score = 71.6 bits (174), Expect = 2e-12
Identities = 34/92 (36%), Positives = 56/92 (59%)
Frame = +2
Query: 127 TRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLRSAGLQT 186
++ + ++ LGC L FI Q D TY N+E +PK+C L++E G+ L ++ LQ
Sbjct: 38 SKDETIQTLGCQCLSKFIYCQVDATYTHNIEKLVPKVCMLSREHGEACEKRCLSASSLQC 217
Query: 187 LSSMVKFMGEHSHLSMDFDKIISAILENYVDL 218
LS+MV FM E SH+ +DFD++ I + +++
Sbjct: 218 LSAMVWFMAEFSHIFVDFDEVCYKIKDAPIEI 313
>BE800242
Length = 247
Score = 67.0 bits (162), Expect = 4e-11
Identities = 40/77 (51%), Positives = 52/77 (66%)
Frame = +3
Query: 635 HTLIALLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSAR 694
+TL+ L+ Y LVR FQLAFSL +ISL +EG LPPSRRRSL TLA+ M++FS++
Sbjct: 12 YTLVLLVSREKNSFYEVLVRSFQLAFSLWNISL-KEGPLPPSRRRSLFTLATSMIVFSSK 188
Query: 695 AADFSDLIPKVKASLTE 711
+ L+ KA LTE
Sbjct: 189 *YNIDHLVQSAKAVLTE 239
>TC227371 similar to GB|AAQ89612.1|37201994|BT010590 At5g26850 {Arabidopsis
thaliana;} , partial (11%)
Length = 1221
Score = 65.1 bits (157), Expect = 1e-10
Identities = 48/157 (30%), Positives = 78/157 (49%), Gaps = 1/157 (0%)
Frame = +1
Query: 652 LVRCFQLAFSLRSISLDQEGG-LPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKASLT 710
++R FQL SL ++ LDQ G L P+ +RS+ L++ ML F+ + DL V ASL
Sbjct: 37 VIRFFQLPLSLWTMLLDQSNGILSPACQRSVYVLSAGMLAFACKIYQIHDL-NDVFASLP 213
Query: 711 EAPVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFM 770
+ VDPFL + DD + A I D +G+ D A LS +Q R+ + + +
Sbjct: 214 MSDVDPFLSISDDYQVYAK-IHVDVREYGTAADNQLACSMLSELQNKIRECQSIIRDALV 390
Query: 771 TKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFME 807
+ + E + + L + F PD+ + GP ++
Sbjct: 391 HNLANVTELDADELAMLLSEKFKPDEEFVFGPQSMLD 501
Score = 32.3 bits (72), Expect = 1.0
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +1
Query: 935 LRLPPSSPYDKFLKAAG 951
++LPP+SP+D FLKAAG
Sbjct: 940 MKLPPASPFDNFLKAAG 990
>BI094993
Length = 350
Score = 51.6 bits (122), Expect(2) = 8e-10
Identities = 23/31 (74%), Positives = 24/31 (77%)
Frame = +1
Query: 791 GFSPDDAYPSGPPLFMETPRPGSPLAQIEFP 821
G PDD+YPSGPP FMETPRP S A IEFP
Sbjct: 256 GLFPDDSYPSGPPFFMETPRPCSYFA*IEFP 348
Score = 30.8 bits (68), Expect(2) = 8e-10
Identities = 13/17 (76%), Positives = 16/17 (93%)
Frame = +2
Query: 778 EDELSSIKNQLLQGFSP 794
+DEL SIKN+LLQG+SP
Sbjct: 218 QDELFSIKNKLLQGYSP 268
>CD408859 weakly similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (4%)
Length = 464
Score = 55.8 bits (133), Expect = 9e-08
Identities = 22/40 (55%), Positives = 33/40 (82%)
Frame = -3
Query: 70 KITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQI 109
+I + LE RCYK+L+NE+F S K+++CIY+K L SC+EQ+
Sbjct: 330 RIVQALEPRCYKELQNENFISTKIVMCIYKKFLFSCKEQM 211
>BM094146 similar to GP|14194169|gb| At1g05960/T21E18_20 {Arabidopsis
thaliana}, partial (6%)
Length = 398
Score = 53.5 bits (127), Expect = 4e-07
Identities = 24/37 (64%), Positives = 29/37 (77%)
Frame = +3
Query: 42 PRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQR 78
PRN+V NDR IGKLC+Y ++NPL IPKI + LEQR
Sbjct: 42 PRNQVEVPNDRSIGKLCDYTARNPLCIPKIVQALEQR 152
Score = 39.7 bits (91), Expect = 0.006
Identities = 27/81 (33%), Positives = 38/81 (46%)
Frame = +2
Query: 29 PVKRYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQRCYKDLRNESF 88
PVKRYKKLIA+I P K K+ + PK + +
Sbjct: 2 PVKRYKKLIADIFPS*PGGSAK**KHWKIM*LYCQKSFMHPKDCASP*AKVIT------- 160
Query: 89 GSVKVILCIYRKLLSSCREQI 109
S K+++CIY+K L SC+EQ+
Sbjct: 161 -STKIVMCIYKKFLFSCKEQM 220
>TC227372 similar to GB|AAQ89612.1|37201994|BT010590 At5g26850 {Arabidopsis
thaliana;} , partial (10%)
Length = 1211
Score = 32.3 bits (72), Expect = 1.0
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +3
Query: 935 LRLPPSSPYDKFLKAAG 951
++LPP+SP+D FLKAAG
Sbjct: 477 MKLPPASPFDNFLKAAG 527
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,394,877
Number of Sequences: 63676
Number of extensions: 530618
Number of successful extensions: 3862
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 3793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3852
length of query: 951
length of database: 12,639,632
effective HSP length: 106
effective length of query: 845
effective length of database: 5,889,976
effective search space: 4977029720
effective search space used: 4977029720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146574.12


