

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
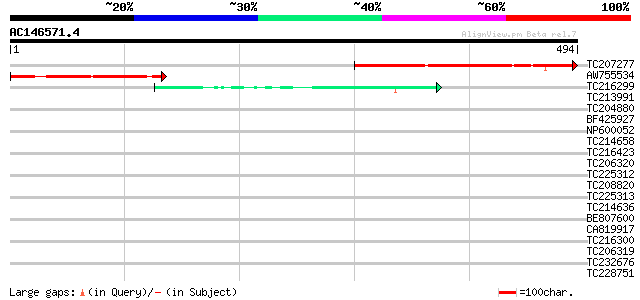
Query= AC146571.4 + phase: 0
(494 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207277 weakly similar to UP|Q7CV43 (Q7CV43) AGR_L_732p, partia... 267 7e-72
AW755534 133 2e-31
TC216299 similar to UP|Q9SSQ3 (Q9SSQ3) F6D8.27 protein, partial ... 50 3e-06
TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial ... 39 0.007
TC204880 similar to UP|Q93ZN4 (Q93ZN4) AT4g12830/T20K18_180, par... 37 0.015
BF425927 37 0.026
NP600052 zeatin O-glucosyltransferase [Glycine max] 35 0.059
TC214658 homologue to UP|Q39856 (Q39856) Epoxide hydrolase, part... 35 0.059
TC216423 UP|O49857 (O49857) Epoxide hydrolase, complete 35 0.077
TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%) 35 0.10
TC225312 similar to UP|Q76E11 (Q76E11) Soluble epoxide hydrolase... 34 0.13
TC208820 similar to UP|Q6NL07 (Q6NL07) At1g13820, partial (83%) 34 0.13
TC225313 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 34 0.13
TC214636 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 34 0.17
BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata},... 34 0.17
CA819917 33 0.22
TC216300 similar to UP|Q9SSQ3 (Q9SSQ3) F6D8.27 protein, partial ... 33 0.22
TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein R... 33 0.29
TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like,... 33 0.38
TC228751 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {A... 33 0.38
>TC207277 weakly similar to UP|Q7CV43 (Q7CV43) AGR_L_732p, partial (6%)
Length = 980
Score = 267 bits (683), Expect = 7e-72
Identities = 134/198 (67%), Positives = 155/198 (77%), Gaps = 4/198 (2%)
Frame = +3
Query: 301 IREVVLGFPYVFAKVVNFYCSKRIGGLDADAHRVLLKSGDGRKAVVAIGKNLNSSFDLTE 360
+REVVLG +VFAKVV CSKR+G D++A R LLK DGR+AVV +GK +NSSF L E
Sbjct: 3 VREVVLGVSFVFAKVVALCCSKRVGVADSEASRALLKGRDGRRAVVNVGKRVNSSFGLEE 182
Query: 361 WGCSDRLKDMPMQLIWSSDWSEEWSSEGNRVAGALPRAKFVTHSGGRWAQEDVAVEIAEK 420
WG + LK MPMQ++WS+ WSEEWS EG+RVA ALP+A FVTH+GGRWAQED AVEIAEK
Sbjct: 183 WG--EGLKGMPMQVMWSAGWSEEWSQEGHRVADALPQASFVTHTGGRWAQEDAAVEIAEK 356
Query: 421 ISQFVSSLPKTVRKVEQEPPIPDHVQKMFDEAKSDDGHDHHQGHS----HGHDRLGEAHI 476
ISQFV SLPK+VRKVEQE IP+H+QKM DEAKS GHDHH HS HGHD G+AHI
Sbjct: 357 ISQFVLSLPKSVRKVEQE-SIPEHIQKMLDEAKS-SGHDHHHHHSHDHGHGHDHYGDAHI 530
Query: 477 HEAGYMDAYGIGHEPNDW 494
H A YMDAYG+GH + W
Sbjct: 531 HGANYMDAYGLGHGHHGW 584
>AW755534
Length = 434
Score = 133 bits (335), Expect = 2e-31
Identities = 74/137 (54%), Positives = 87/137 (63%), Gaps = 1/137 (0%)
Frame = +1
Query: 1 MAIITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTN-PLSFWFYFTLSI 59
MAIITEE E++ Q K + K K + PS+ P N P SFWFYFTLS+
Sbjct: 61 MAIITEEPEAEADSHSQIPKPR---------KPKLKTPSSPPPNSNNNPFSFWFYFTLSV 213
Query: 60 SLVTLFFLFTSSPLSSSSPQQWFLTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDP 119
SL+TLFF+ TSS LS P+ WFLTLP LR HYSNGRTIKVQ H N+ PI +FT Q P
Sbjct: 214 SLLTLFFVLTSS-LSPQDPKTWFLTLPPPLRHHYSNGRTIKVQTHSNEAPIQVFTLQEGP 390
Query: 120 TIPNPSETVLILHGQAL 136
T SE ++ILHGQ L
Sbjct: 391 T---SSENIIILHGQGL 432
>TC216299 similar to UP|Q9SSQ3 (Q9SSQ3) F6D8.27 protein, partial (84%)
Length = 1455
Score = 49.7 bits (117), Expect = 3e-06
Identities = 66/253 (26%), Positives = 97/253 (38%), Gaps = 3/253 (1%)
Frame = +3
Query: 127 TVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVSVEGLDGIFGRFSY 186
T++ LHG S+SYR ++ LS G A D G GFSDK +G F+Y
Sbjct: 444 TIIFLHGAPTQSFSYRVVMSQLSDAGFHCFAPDWIGFGFSDKP--------QPGYG-FNY 596
Query: 187 VYSEIKEKGFFWAFDQIVETGQIPYEEVLARMSKRKVNKPIDLGPEEIGKVLGEVIGTLG 246
EK F A D+++ EVL V P L V G ++G+ G
Sbjct: 597 T-----EKEFHDALDKLL--------EVLG------VKSPFFL------VVQGFLVGSYG 701
Query: 247 LAPVHLVLHDSALGFTANWVSENSDLVSSLTLVDTPVPPSNLGAFPIWVLDVPLIREVVL 306
L W +NS +S L ++++P+ S+ L +PL E
Sbjct: 702 L----------------TWALKNSSKISKLAILNSPLTDSSPIPGLFQKLRIPLYGEFTS 833
Query: 307 GFPYVFAKVVNFYCSKRIGGLDADAHRV--LLKSGDGRKAVVAIGK-NLNSSFDLTEWGC 363
+ + + + AD +R+ L SG G + A K N +F G
Sbjct: 834 QNAIIAERFIEGGSPYVLKNEKADVYRLPYLASSGPGFALLEAARKANFKGTFSEISAGF 1013
Query: 364 SDRLKDMPMQLIW 376
+ D PM L W
Sbjct: 1014TTERWDKPMLLAW 1052
>TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial (12%)
Length = 440
Score = 38.5 bits (88), Expect = 0.007
Identities = 18/39 (46%), Positives = 29/39 (74%)
Frame = +3
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
+Q+ QQQQQQ+Q+Q+Q+QQ + +S+ Q+ +QPQQ
Sbjct: 3 QQQQQQQQQQQQQQQQQQQQQQQQPQSQQQQ---SQPQQ 110
Score = 37.7 bits (86), Expect = 0.012
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = +3
Query: 8 QESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQT 46
Q+ QQQQQQ+Q+Q+Q+QQ + Q+ S Q QQ+
Sbjct: 3 QQQQQQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQS 119
Score = 36.2 bits (82), Expect = 0.034
Identities = 14/37 (37%), Positives = 26/37 (69%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQ 42
++Q+ QQQQQQ+Q+Q+Q+QQ +++ + +P Q
Sbjct: 6 QQQQQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQ 116
Score = 32.0 bits (71), Expect = 0.65
Identities = 14/42 (33%), Positives = 24/42 (56%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTN 47
++Q+ QQQQQQ+Q+Q+Q+ Q + Q+ S + N
Sbjct: 18 QQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQSRDRAHLLN 143
>TC204880 similar to UP|Q93ZN4 (Q93ZN4) AT4g12830/T20K18_180, partial (83%)
Length = 1213
Score = 37.4 bits (85), Expect = 0.015
Identities = 22/46 (47%), Positives = 29/46 (62%)
Frame = +1
Query: 123 NPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDK 168
NPS V+++HG +YSYR ++ LS + VIA D G GFSDK
Sbjct: 208 NPS--VILIHGFPSQAYSYRKVLPVLS-KDYHVIAFDWLGFGFSDK 336
>BF425927
Length = 427
Score = 36.6 bits (83), Expect = 0.026
Identities = 32/132 (24%), Positives = 57/132 (42%), Gaps = 8/132 (6%)
Frame = +1
Query: 22 KQKQQHSNKTKSKTQKPSTTQPQQTNP--LSFWFYFTLSISLVTLFFLFTSSPLSSSSPQ 79
+++QQH +S++ Q P L + F S +L++ FF+F SP SSSSP
Sbjct: 40 REEQQHGRDDRSRSSWAEVVSQDQQVPFQLPCFLIFASSPNLISSFFIFAGSP-SSSSPT 216
Query: 80 QWFLTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQL------DPTIPNPSETVLILHG 133
Q ++ ++ I P Q + L +F L D + + + ++H
Sbjct: 217 QGLIS------------SSLLFSIPPKQTCVILSSFSL*EEQCQDYSYRHRPHKLNLIHF 360
Query: 134 QALSSYSYRNLI 145
+ S+ Y N +
Sbjct: 361 SSTSNVKYYNTL 396
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 35.4 bits (80), Expect = 0.059
Identities = 30/79 (37%), Positives = 35/79 (43%), Gaps = 8/79 (10%)
Frame = +1
Query: 88 TLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDPTIPNPSETVLILHGQALSSYS------- 140
TLR H SN I + H + P F P PN ET H L S+
Sbjct: 163 TLRDHNSNISNIIIHFHAFEVP----PFVSPPPNPNNEETDFPSH--LLPSFKASSHLRE 324
Query: 141 -YRNLIQSLSTQGVRVIAI 158
RNL+QSLS+Q RVI I
Sbjct: 325 PVRNLLQSLSSQAKRVIVI 381
>TC214658 homologue to UP|Q39856 (Q39856) Epoxide hydrolase, partial (87%)
Length = 1085
Score = 35.4 bits (80), Expect = 0.059
Identities = 21/46 (45%), Positives = 27/46 (58%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG YS+R+ I SLS+ G R +A DL G G ++ VS
Sbjct: 94 VLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLRGYGDTEAPPSVS 231
>TC216423 UP|O49857 (O49857) Epoxide hydrolase, complete
Length = 1343
Score = 35.0 bits (79), Expect = 0.077
Identities = 20/46 (43%), Positives = 27/46 (58%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG YS+R+ I SLS+ G R +A DL G G ++ +S
Sbjct: 226 VLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLRGYGDTEAPPSIS 363
>TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%)
Length = 1120
Score = 34.7 bits (78), Expect = 0.10
Identities = 17/39 (43%), Positives = 22/39 (55%)
Frame = +2
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
+Q QQQQQQ Q+Q Q QQH + + Q+ TT Q
Sbjct: 362 QQPQPQQQQQQHQQQLQLQQHMQQQLQQQQQQETTSQLQ 478
Score = 31.2 bits (69), Expect = 1.1
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = +2
Query: 9 ESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
+ QQQ QQ Q Q+Q+QQH + + + Q QQ
Sbjct: 341 QQQQQQHQQPQPQQQQQQHQQQLQLQQHMQQQLQQQQ 451
>TC225312 similar to UP|Q76E11 (Q76E11) Soluble epoxide hydrolase, partial
(96%)
Length = 1213
Score = 34.3 bits (77), Expect = 0.13
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = +2
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG Y++R+ + SLS G R IA DL G G +D + S
Sbjct: 119 VLFLHGFPELWYTWRHQLLSLSAVGYRAIAPDLRGYGDTDAPPDAS 256
>TC208820 similar to UP|Q6NL07 (Q6NL07) At1g13820, partial (83%)
Length = 1422
Score = 34.3 bits (77), Expect = 0.13
Identities = 17/49 (34%), Positives = 25/49 (50%)
Frame = +2
Query: 119 PTIPNPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSD 167
P + V++LHG S +R ++ L G+ AID+ G GFSD
Sbjct: 428 PLVQTKETPVVLLHGFDSSCLEWRYVLPLLEESGIETWAIDILGWGFSD 574
>TC225313 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (51%)
Length = 1307
Score = 34.3 bits (77), Expect = 0.13
Identities = 19/46 (41%), Positives = 26/46 (56%)
Frame = +3
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG YS+R+ I +LS G R +A DL G G ++ +S
Sbjct: 705 VLFLHGFPELWYSWRHQILALSNLGYRAVAPDLRGYGDTEAPASIS 842
>TC214636 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (58%)
Length = 778
Score = 33.9 bits (76), Expect = 0.17
Identities = 19/37 (51%), Positives = 23/37 (61%)
Frame = +3
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSG 164
VL LHG YS+R+ I SLS+ G R +A DL G G
Sbjct: 138 VLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLRGYG 248
>BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata}, partial
(4%)
Length = 414
Score = 33.9 bits (76), Expect = 0.17
Identities = 13/24 (54%), Positives = 20/24 (83%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSN 29
++Q+ QQQQQQ+Q+Q+Q+QQ N
Sbjct: 336 QQQQQQQQQQQQQQQQQQQQQQQN 407
Score = 32.3 bits (72), Expect = 0.50
Identities = 12/25 (48%), Positives = 21/25 (84%)
Frame = +3
Query: 4 ITEEQESDQQQQQQKQKQKQKQQHS 28
+ ++Q+ QQQQQQ+Q+Q+Q+QQ +
Sbjct: 333 LQQQQQQQQQQQQQQQQQQQQQQQN 407
Score = 30.4 bits (67), Expect = 1.9
Identities = 12/23 (52%), Positives = 18/23 (78%)
Frame = +3
Query: 8 QESDQQQQQQKQKQKQKQQHSNK 30
Q+ QQQQQQ+Q+Q+Q+QQ +
Sbjct: 336 QQQQQQQQQQQQQQQQQQQQQQQ 404
Score = 30.0 bits (66), Expect = 2.5
Identities = 11/28 (39%), Positives = 21/28 (74%)
Frame = +3
Query: 3 IITEEQESDQQQQQQKQKQKQKQQHSNK 30
++ ++ + QQQQQQ+Q+Q+Q+QQ +
Sbjct: 318 LLRQKLQQQQQQQQQQQQQQQQQQQQQQ 401
Score = 28.5 bits (62), Expect = 7.2
Identities = 11/30 (36%), Positives = 21/30 (69%)
Frame = +3
Query: 4 ITEEQESDQQQQQQKQKQKQKQQHSNKTKS 33
+ ++ QQQQQQ+Q+Q+Q+QQ + ++
Sbjct: 318 LLRQKLQQQQQQQQQQQQQQQQQQQQQQQN 407
Score = 28.1 bits (61), Expect = 9.4
Identities = 11/23 (47%), Positives = 18/23 (77%)
Frame = +3
Query: 12 QQQQQQKQKQKQKQQHSNKTKSK 34
QQQQQQ+Q+Q+Q+QQ + + +
Sbjct: 336 QQQQQQQQQQQQQQQQQQQQQQQ 404
>CA819917
Length = 418
Score = 33.5 bits (75), Expect = 0.22
Identities = 23/82 (28%), Positives = 42/82 (51%)
Frame = +1
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLSFWFYFTLSISLVTLF 65
++++ ++++++K+K+K+K++ K K K +KP PQ L F+F+F F
Sbjct: 4 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKPGGGPPQIF--LLFFFFF-----FCPFF 162
Query: 66 FLFTSSPLSSSSPQQWFLTLPT 87
F F P +FLT T
Sbjct: 163FAFFLKGKGEKKP--FFLTFLT 222
>TC216300 similar to UP|Q9SSQ3 (Q9SSQ3) F6D8.27 protein, partial (46%)
Length = 777
Score = 33.5 bits (75), Expect = 0.22
Identities = 17/42 (40%), Positives = 22/42 (51%)
Frame = +1
Query: 127 TVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDK 168
T+ LHG S+SYR ++ L+ G A D G GF DK
Sbjct: 436 TIDFLHGAPTQSFSYRIVMSQLTDAGFHCFAPDWIGLGFIDK 561
>TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein RIPA, partial
(5%)
Length = 1428
Score = 33.1 bits (74), Expect = 0.29
Identities = 16/45 (35%), Positives = 25/45 (55%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
++ + QQQQQQ Q+Q Q QQH + + Q+ T Q + +S
Sbjct: 693 QQHQQPQQQQQQHQQQLQLQQHMQQQLQQQQQQQETTSQLQSVVS 827
Score = 29.3 bits (64), Expect = 4.2
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +3
Query: 13 QQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQ 45
QQQQ +Q Q+Q+QQH + + + Q QQ
Sbjct: 687 QQQQHQQPQQQQQQHQQQLQLQQHMQQQLQQQQ 785
>TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like, partial
(57%)
Length = 926
Score = 32.7 bits (73), Expect = 0.38
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = +1
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQP 43
E Q QQQQQQ+Q+Q+Q+QQ S + P P
Sbjct: 490 EMQVQQQQQQQQQQQQQQQQQQSQGLGEQVSMPMYNLP 603
Score = 32.0 bits (71), Expect = 0.65
Identities = 13/25 (52%), Positives = 20/25 (80%)
Frame = +1
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNK 30
EE + QQQQQQ+Q+Q+Q+QQ ++
Sbjct: 487 EEMQVQQQQQQQQQQQQQQQQQQSQ 561
Score = 30.0 bits (66), Expect = 2.5
Identities = 13/21 (61%), Positives = 18/21 (84%)
Frame = +1
Query: 6 EEQESDQQQQQQKQKQKQKQQ 26
EE+ QQQQQQ+Q+Q+Q+QQ
Sbjct: 484 EEEMQVQQQQQQQQQQQQQQQ 546
Score = 28.1 bits (61), Expect = 9.4
Identities = 11/23 (47%), Positives = 17/23 (73%)
Frame = +1
Query: 1 MAIITEEQESDQQQQQQKQKQKQ 23
M + ++Q+ QQQQQQ+Q+Q Q
Sbjct: 493 MQVQQQQQQQQQQQQQQQQQQSQ 561
>TC228751 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {Arabidopsis
thaliana;} , partial (81%)
Length = 1381
Score = 32.7 bits (73), Expect = 0.38
Identities = 19/46 (41%), Positives = 28/46 (60%)
Frame = +2
Query: 123 NPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDK 168
N ++++HG +S +RN I L+ Q RV +IDL G G+SDK
Sbjct: 296 NIGPALVLVHGFGANSDHWRNNISVLA-QSHRVYSIDLIGYGYSDK 430
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,208,838
Number of Sequences: 63676
Number of extensions: 373391
Number of successful extensions: 3773
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 3211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3490
length of query: 494
length of database: 12,639,632
effective HSP length: 101
effective length of query: 393
effective length of database: 6,208,356
effective search space: 2439883908
effective search space used: 2439883908
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146571.4


