

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.1 - phase: 0
(713 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
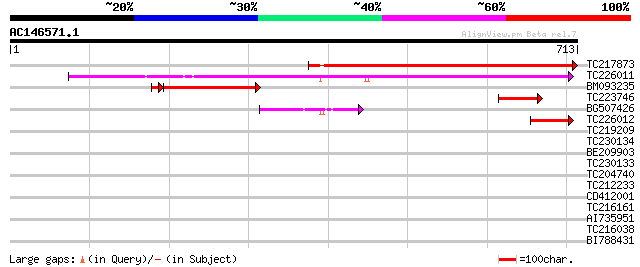
Score E
Sequences producing significant alignments: (bits) Value
TC217873 similar to UP|Q8RY11 (Q8RY11) AT3g05350/T12H1_32, parti... 625 e-179
TC226011 similar to UP|Q93X45 (Q93X45) Xaa-Pro aminopeptidase 2 ... 516 e-146
BM093235 similar to GP|19310478|gb AT3g05350/T12H1_32 {Arabidops... 219 7e-60
TC223746 similar to UP|Q93X45 (Q93X45) Xaa-Pro aminopeptidase 2 ... 58 1e-08
BG507426 similar to GP|15384989|emb Xaa-Pro aminopeptidase 1 {Ly... 58 2e-08
TC226012 similar to UP|O23206 (O23206) Aminopeptidase-like prote... 52 9e-07
TC219209 similar to GB|BAA95736.1|7939533|AB025608 Nicotiana ERE... 33 0.34
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 33 0.57
BE209903 similar to GP|8809606|dbj| ethylene responsive element ... 31 1.7
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 31 2.2
TC204740 similar to UP|Q40478 (Q40478) Ethylene-responsive eleme... 30 2.8
TC212233 30 2.8
CD412001 30 3.7
TC216161 similar to UP|NO93_SOYBN (Q02921) Early nodulin 93 (N-9... 30 4.8
AI735951 29 8.3
TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, pa... 29 8.3
BI788431 29 8.3
>TC217873 similar to UP|Q8RY11 (Q8RY11) AT3g05350/T12H1_32, partial (42%)
Length = 1231
Score = 625 bits (1613), Expect = e-179
Identities = 302/338 (89%), Positives = 318/338 (93%)
Frame = +2
Query: 376 NYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQ 435
N E+KHKTR+ GFDGS SDVP +VHK SPVS AKAIKNE+EL+GM+ CHLRDAAALAQ
Sbjct: 2 NCENKHKTRTNGFDGS---SDVPFSVHKVSPVSQAKAIKNESELEGMRNCHLRDAAALAQ 172
Query: 436 FWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGS 495
FWDWLE EIT DRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEP S
Sbjct: 173 FWDWLETEITKDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPES 352
Query: 496 CSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPED 555
CS+VDANKLFLLDSGAQYVDGTTDITRTVHFGKPT REKECFTRVLQGHIALDQ+VFPE+
Sbjct: 353 CSSVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTAREKECFTRVLQGHIALDQSVFPEN 532
Query: 556 TPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSN 615
TPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQ IS+RYGNLTPLV GMIVSN
Sbjct: 533 TPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQSISHRYGNLTPLVKGMIVSN 712
Query: 616 EPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEI 675
EPGYYEDHAFGIRIENLLYVRN ETPNRFGGI+YLGFEKLTYVPIQIKLVD+SLLS EI
Sbjct: 713 EPGYYEDHAFGIRIENLLYVRNAETPNRFGGIEYLGFEKLTYVPIQIKLVDLSLLSAAEI 892
Query: 676 DWLNNYHSVVWEKVSPLLDGSARQWLWNNTQPIIRETV 713
DWLNNYHS+VWEKVSPL+DGSARQWLWNNT+PII E +
Sbjct: 893 DWLNNYHSLVWEKVSPLMDGSARQWLWNNTRPIIHEKI 1006
>TC226011 similar to UP|Q93X45 (Q93X45) Xaa-Pro aminopeptidase 2 , partial
(94%)
Length = 2304
Score = 516 bits (1328), Expect = e-146
Identities = 285/667 (42%), Positives = 399/667 (59%), Gaps = 33/667 (4%)
Frame = +3
Query: 75 RSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGT 134
++ + + ++ALR L +DA ++PS+D H SE+++ RR+++S FTGS G
Sbjct: 33 KTEENKMEDTVSALRSLMVSHSPPLDALVVPSEDYHLSEYVSARDKRREFVSGFTGSAGL 212
Query: 135 AVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPF 194
A++T +A LWTDGRYFLQAEK+L++ W LMR G P W+ + L A VG+DP+
Sbjct: 213 ALITKKEALLWTDGRYFLQAEKELSAGWKLMRIGED--PAVDIWMADNLPKEASVGVDPW 386
Query: 195 LFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVAS 254
+ D A+ + ++K +LV + NLVDE+W RP V VH +K+AG VA
Sbjct: 387 CISIDTAQRWERAFAEKQQKLVPT-SKNLVDEVWIN-RPPAEINAVIVHPVKFAGRSVAD 560
Query: 255 KLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNS 314
KL LR +LV + II TALDE+AWL N+RGSD+ + PVV+A+ IV + A +++D
Sbjct: 561 KLKDLRKKLVHEQTRGIIFTALDEVAWLYNIRGSDVAYCPVVHAFAIVTSNSAFIYVDKQ 740
Query: 315 KVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYY 374
KV+ EV HL + IEI+ Y ++ S+ LA + T+ A A K +
Sbjct: 741 KVSVEVQTHLVENGIEIQEYTAVSSDATLLATDELDA-VSTAKAALAETEARKIPSE-ID 914
Query: 375 QNYESKHKTRSKGFD-----------GSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQ 423
++ +H+ D A + + + SP++LAKA+KN EL G++
Sbjct: 915 KSVNGEHQAEENSNDLIWADPVSCCYALYAKLNPDTVLLQQSPLALAKALKNSVELDGLK 1094
Query: 424 ECHLRDAAALAQFWDWLEKEITN-----------DRI----------LTEVEVSDKLLEF 462
+ H+RD AA+ Q+ WL+K++ + D + LTEV VSD+L F
Sbjct: 1095KAHIRDGAAVVQYLVWLDKKMQDIYGASGYFLEKDSVKKEKHLQSLKLTEVTVSDQLEGF 1274
Query: 463 RSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITR 522
R+ + F SF TIS G N AIIHY P+ +C+ +D +K++L DSGAQY+DGTTDITR
Sbjct: 1275RASKEHFKGLSFPTISSVGSNAAIIHYFPKAETCAELDPDKIYLFDSGAQYLDGTTDITR 1454
Query: 523 TVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGH 582
TVHFGKP+T EK C+T VL+GHIAL A FP T G LD AR LWK GLDYRHGTGH
Sbjct: 1455TVHFGKPSTHEKACYTAVLKGHIALGNARFPNGTNGHSLDILARIPLWKDGLDYRHGTGH 1634
Query: 583 GVGAALNVHEGPQGISYR-YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETP 641
G+G+ LNVHEGP IS+R PL + M V++EPGYYED FGIR+EN+L V+ +T
Sbjct: 1635GIGSYLNVHEGPHLISFRPQARNVPLQSSMTVTDEPGYYEDGEFGIRLENVLIVKEADTT 1814
Query: 642 NRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWL 701
FG YL FE +T+ P Q KL+D++LLS EI+WLN+YH+ ++P LD WL
Sbjct: 1815FNFGDRGYLSFEHITWAPYQTKLIDLNLLSPEEINWLNSYHATCRNILAPYLDEVENAWL 1994
Query: 702 WNNTQPI 708
T+P+
Sbjct: 1995KKATEPV 2015
>BM093235 similar to GP|19310478|gb AT3g05350/T12H1_32 {Arabidopsis
thaliana}, partial (19%)
Length = 420
Score = 219 bits (557), Expect(2) = 7e-60
Identities = 108/122 (88%), Positives = 115/122 (93%)
Frame = +3
Query: 194 FLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVA 253
FLFTSDAAEELK VISK NHELVYLYNSNLVDEIWKE+RP+PPN PVRVH LKYAGLDVA
Sbjct: 54 FLFTSDAAEELKGVISKNNHELVYLYNSNLVDEIWKESRPKPPNNPVRVHNLKYAGLDVA 233
Query: 254 SKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDN 313
SKLSSLRSELV AGSSAI+++ LDEIAWLLNLRGSDIPHSPVVYAYLIVE+ GAKLFID+
Sbjct: 234 SKLSSLRSELVNAGSSAIVISMLDEIAWLLNLRGSDIPHSPVVYAYLIVEVSGAKLFIDD 413
Query: 314 SK 315
SK
Sbjct: 414 SK 419
Score = 31.2 bits (69), Expect(2) = 7e-60
Identities = 13/15 (86%), Positives = 14/15 (92%)
Frame = +2
Query: 179 LNEVLAPGARVGIDP 193
LN+VLAPG RVGIDP
Sbjct: 2 LNDVLAPGGRVGIDP 46
>TC223746 similar to UP|Q93X45 (Q93X45) Xaa-Pro aminopeptidase 2 , partial
(6%)
Length = 443
Score = 58.2 bits (139), Expect = 1e-08
Identities = 26/56 (46%), Positives = 38/56 (67%)
Frame = +1
Query: 615 NEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLL 670
+EPGYYED FGIR+EN+L V+ +T FG YL FE +T+V + ++ +SL+
Sbjct: 199 SEPGYYEDGEFGIRLENVLIVKEADTTFNFGDRGYLSFEHITWVSSSMFIISLSLI 366
>BG507426 similar to GP|15384989|emb Xaa-Pro aminopeptidase 1 {Lycopersicon
esculentum}, partial (11%)
Length = 436
Score = 57.8 bits (138), Expect = 2e-08
Identities = 45/143 (31%), Positives = 72/143 (49%), Gaps = 13/143 (9%)
Frame = +3
Query: 315 KVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYY 374
KV+ EV HL + IEIR Y ++ S+ LA + T+ V A K +
Sbjct: 3 KVSVEVQAHLVENGIEIREYTAVSSDTTLLATDELDS-VSTAKVALAETEVRKIP-NETA 176
Query: 375 QNYESKHKTRSKGFD------GSIA-------NSDVPIAVHKSSPVSLAKAIKNETELKG 421
++ +H+ D GS N D + +H+S P++LAKA+KN EL G
Sbjct: 177 KHANGEHQAEENSNDLIWADPGSCCYALYAKLNPDT-VLLHQS-PLALAKALKNSVELDG 350
Query: 422 MQECHLRDAAALAQFWDWLEKEI 444
+++ H+RD AA+ Q+ WL+K++
Sbjct: 351 LKKAHIRDGAAVVQYLVWLDKKM 419
>TC226012 similar to UP|O23206 (O23206) Aminopeptidase-like protein, partial
(8%)
Length = 852
Score = 52.0 bits (123), Expect = 9e-07
Identities = 23/54 (42%), Positives = 33/54 (60%)
Frame = +2
Query: 655 LTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNTQPI 708
L P Q KL+D++LL EIDWLN+YHS + ++P L+ WL T+P+
Sbjct: 470 LVQAPYQTKLIDLNLLCPEEIDWLNSYHSTCRDILAPYLNEVENAWLKKATEPV 631
>TC219209 similar to GB|BAA95736.1|7939533|AB025608 Nicotiana EREBP-3-like
protein {Arabidopsis thaliana;} , partial (45%)
Length = 781
Score = 33.5 bits (75), Expect = 0.34
Identities = 24/115 (20%), Positives = 50/115 (42%), Gaps = 5/115 (4%)
Frame = +2
Query: 303 EIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAI 362
+++G + + N V ++ I RP+ +EI + +G+ +WL T
Sbjct: 95 KMEGGRSSVSNGNV------EVRYRGIRRRPWGKFAAEIRDPTRKGTRIWLGTFDTAEQA 256
Query: 363 VNAYKAAC-----DRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKA 412
AY AA R N+ +++++ + NS +P+ + S+P S + +
Sbjct: 257 ARAYDAAAFHFRGHRAILNFPNEYQSHN-------PNSSLPMPLAVSAPPSYSSS 400
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 32.7 bits (73), Expect = 0.57
Identities = 31/89 (34%), Positives = 47/89 (51%)
Frame = -3
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSIS 64
S SSSSSSSP++ + + S SS + +L+P S++ P S ++S+S S
Sbjct: 507 SSSSSSSSSPSSSSLSSSSSSSSSSSSSLSPPPLASGSSSESASSPPSSPEKSSSSSS-S 331
Query: 65 AKPSSQLRKNRSSNSDSDPKLTALRRLFS 93
+ PSS + SS+S S +L LFS
Sbjct: 330 SSPSSPSFTSSSSSSSS----VSLASLFS 256
Score = 28.9 bits (63), Expect = 8.3
Identities = 27/79 (34%), Positives = 42/79 (52%)
Frame = -3
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSIS 64
S SSSSSSS ++ + +LS SS + + S+ + PP + + ++S S S
Sbjct: 516 SSSSSSSSSSSSPSSSSLSSSSSSSSSS----------SSSLSPPP---LASGSSSESAS 376
Query: 65 AKPSSQLRKNRSSNSDSDP 83
+ PSS K+ SS+S S P
Sbjct: 375 SPPSSP-EKSSSSSSSSSP 322
>BE209903 similar to GP|8809606|dbj| ethylene responsive element binding
factor 5 (ATERF5) {Arabidopsis thaliana}, partial (16%)
Length = 175
Score = 31.2 bits (69), Expect = 1.7
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +2
Query: 332 RPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAY 366
RP+ +EI + RGS +WL T + VNAY
Sbjct: 20 RPWGKXAAEIRDPNKRGSRVWLRTLDTDGEAVNAY 124
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 30.8 bits (68), Expect = 2.2
Identities = 28/85 (32%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Frame = -1
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKH-NKPPFFSIRNCTTSNSI 63
S SSSSSS + + L+ S SS + +L P S++ + PP ++ ++S+S+
Sbjct: 508 SSPSSSSSSSPSSSSLSSSSSSSPSSSSLPPPPLASGSSSESASSPPSSPEKSSSSSSSL 329
Query: 64 SAKPSSQLRKNRSSNSDSDPKLTAL 88
S+ PS L SS+S S L +L
Sbjct: 328 SSSPSFTL----SSSSSSSVSLASL 266
>TC204740 similar to UP|Q40478 (Q40478) Ethylene-responsive element binding
protein, partial (26%)
Length = 1370
Score = 30.4 bits (67), Expect = 2.8
Identities = 24/106 (22%), Positives = 38/106 (35%)
Frame = +1
Query: 313 NSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDR 372
+S T D + RP+ +EI + +GS +WL T AY A +
Sbjct: 502 SSDETNNTKDDKHYRGVRRRPWGKYAAEIRDPNRKGSRVWLGTFDTAIEAAKAYDKAAFK 681
Query: 373 YYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETE 418
+ + G D S S + SS + + +K E E
Sbjct: 682 MRGSKAILNFPLEIGSDQSKEESSSSSSSVSSSSIVIKVGVKRERE 819
>TC212233
Length = 661
Score = 30.4 bits (67), Expect = 2.8
Identities = 29/112 (25%), Positives = 51/112 (44%)
Frame = +1
Query: 408 SLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQA 467
SL + K ++ G+++C + D I D L+E + SDK LE +
Sbjct: 76 SLVQVAKRRSD-GGVKKCRIHD--------------ILRDLCLSESK-SDKFLEVCT--- 198
Query: 468 GFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTD 519
+++ DT+S + P IH+KP+ S V AN + + ++ G+ D
Sbjct: 199 ---NSNIDTVSDTNPRRMSIHWKPD----SDVSANTFNKSCTRSMFIFGSDD 333
>CD412001
Length = 597
Score = 30.0 bits (66), Expect = 3.7
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 11/58 (18%)
Frame = +2
Query: 26 SSFPTLNLNPIFFYKFKSNK-----------HNKPPFFSIRNCTTSNSISAKPSSQLR 72
S T+N+ PIF S + HN PP FS R +S+S S+ S LR
Sbjct: 92 SKLHTINIEPIF*CHILSERYVPESSSMRFLHNHPPIFSKRTHVSSDSNSSISSGSLR 265
>TC216161 similar to UP|NO93_SOYBN (Q02921) Early nodulin 93 (N-93), partial
(93%)
Length = 534
Score = 29.6 bits (65), Expect = 4.8
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 12/78 (15%)
Frame = -3
Query: 331 IRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYES----------- 379
++P I N RGS + +SS ++V+ Y+ C R+ NYES
Sbjct: 430 VKPNPKSKLSIWNSCIRGSVEGVLSSSCQDSLVSYYEICCSRHCGNYESLSRVIEIGSCP 251
Query: 380 -KHKTRSKGFDGSIANSD 396
KH S+ + GS +S+
Sbjct: 250 RKHPYTSQSWYGSSNSSN 197
>AI735951
Length = 436
Score = 28.9 bits (63), Expect = 8.3
Identities = 28/79 (35%), Positives = 36/79 (45%)
Frame = +3
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIRNCTTSNSIS 64
S +SSSSSSP++ + A S SS P + P S+ PP S T+S S
Sbjct: 195 STTSSSSSSPSSCSWAAPSPSSSTPLTSAGP-----SASHPTPIPPGPSSSGPTSSTS-- 353
Query: 65 AKPSSQLRKNRSSNSDSDP 83
P S RSS+S P
Sbjct: 354 --PKSSNSSTRSSSSSPAP 404
>TC216038 similar to UP|Q9IZF4 (Q9IZF4) Aphid transmission P5, partial (3%)
Length = 1754
Score = 28.9 bits (63), Expect = 8.3
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = -2
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLN 32
S SSS+SSSP++ +LS+ S FP L+
Sbjct: 844 SCSSSTSSSPSSKISSSLSLSSKFPCLS 761
>BI788431
Length = 403
Score = 28.9 bits (63), Expect = 8.3
Identities = 14/32 (43%), Positives = 20/32 (61%)
Frame = -3
Query: 5 SLSSSSSSSPATVTKLALSIHSSFPTLNLNPI 36
++S+S +SSP ++ K L I P NLNPI
Sbjct: 299 TISTSCTSSPRSLQKQILQIGHICPDFNLNPI 204
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,034,368
Number of Sequences: 63676
Number of extensions: 447755
Number of successful extensions: 2524
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 2477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2507
length of query: 713
length of database: 12,639,632
effective HSP length: 104
effective length of query: 609
effective length of database: 6,017,328
effective search space: 3664552752
effective search space used: 3664552752
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146571.1


