

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.8 - phase: 0
(647 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
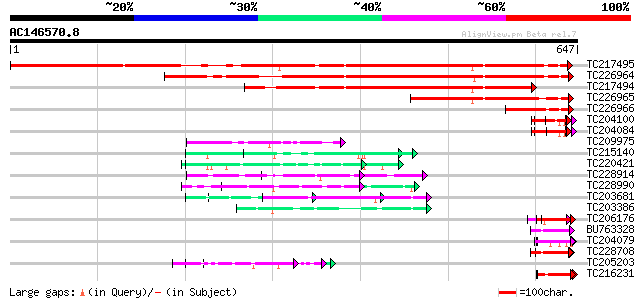
Score E
Sequences producing significant alignments: (bits) Value
TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, part... 788 0.0
TC226964 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 543 e-155
TC217494 weakly similar to UP|Q8BSY2 (Q8BSY2) Mus musculus 11 da... 390 e-108
TC226965 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, part... 276 3e-74
TC226966 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, part... 112 5e-25
TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, ... 57 3e-08
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 57 3e-08
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 56 6e-08
TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 56 6e-08
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 55 1e-07
TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (... 55 1e-07
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 54 2e-07
TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 54 2e-07
TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expr... 54 3e-07
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 52 6e-07
BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall struc... 52 6e-07
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 52 8e-07
TC228708 similar to UP|GRP2_PHAVU (P10496) Glycine-rich cell wal... 52 1e-06
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 52 1e-06
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 50 2e-06
>TC217495 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, partial (59%)
Length = 2299
Score = 788 bits (2036), Expect = 0.0
Identities = 439/674 (65%), Positives = 481/674 (71%), Gaps = 32/674 (4%)
Frame = +2
Query: 1 MASDTASRSSSAADSYIGCLISLTSKSEIRYEGVLYNINTDESSIGLKNVRSFGTEGRKK 60
MAS++ASRSSSAADSYIG LISLTSKSEIRYEG+LYNINT+ESSIGL+NVRSFGTEGRKK
Sbjct: 89 MASESASRSSSAADSYIGSLISLTSKSEIRYEGILYNINTEESSIGLRNVRSFGTEGRKK 268
Query: 61 DGPQILPGDKVYEYILFRGTDIKDLQVKSSPPVQPAVPTNTDPAIIQSQYPRLATTSTSL 120
DGPQI P DKVYEYILFRGTDIKDLQVKSSPPVQP N DPAIIQS YP TTST+L
Sbjct: 269 DGPQIPPSDKVYEYILFRGTDIKDLQVKSSPPVQPTPQVNNDPAIIQSHYPHPITTSTNL 448
Query: 121 P-AVSGSLTDASPNPNTTQLGHPGSNFQGPL--YQPGGNVVSWGASSPAPNANGGGLAMP 177
P AVSGSLTD P+ +TTQ G PGSNFQGPL YQPGGN+ SWGAS PAPNANGG LAMP
Sbjct: 449 PSAVSGSLTD--PSSHTTQHGLPGSNFQGPLPLYQPGGNIGSWGASPPAPNANGGRLAMP 622
Query: 178 -MYWQGYYGAPNGLPQLHQQSLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQ 236
MYWQGYYGAPNGLPQLHQQ SL QP P
Sbjct: 623 PMYWQGYYGAPNGLPQLHQQ---------------------SLLQPPPGLS--------- 712
Query: 237 QPMPSSMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNI 296
MPSSMQQPM Q+P+ + LPT SN PE PS+ LP S+P+I
Sbjct: 713 ----------MPSSMQQPM--------QYPNITPPLPTVSSNLPELPSSLLPASASTPSI 838
Query: 297 TSTSAPPLNL-----------------------STTILPPAPSATLAPETFQASVSNKAP 333
TS S PP NL S++ L PAPSATLA E SV+NKAP
Sbjct: 839 TSASLPPSNLPPAPSALAPAPSALPPAPSALSPSSSALSPAPSATLASENLPVSVTNKAP 1018
Query: 334 TVSLPAATLGANLPSLAPFTSGGSDINAAVP-LANKPNAISGSSLSYQTVPQQMTPSIVG 392
VS AA L ANLPSL T G DINA VP +++KP+AISGSSL YQTV Q +P++VG
Sbjct: 1019NVSTSAAMLAANLPSL---TISGPDINAIVPPISSKPHAISGSSLPYQTV-SQFSPAVVG 1186
Query: 393 SSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVAA 452
SS S+ TE PSL+TP QLLQ G ++ +S++PSQ PHKDVEVVQVSSTS+PEPSVP +A
Sbjct: 1187SSTSIHTETSAPSLLTPGQLLQPGPSIVSSAQPSQAPHKDVEVVQVSSTSSPEPSVPFSA 1366
Query: 453 ESQPPILPLPATSRPIHRPGGASNQTHHGYGYRGRGRGRGIGGFRPAERFTEDFDFTAMN 512
E+QPPILPLP TSRP +RPGGA QTHHGY YRGRGRGRG GG RP +FTEDFDF AMN
Sbjct: 1367ETQPPILPLPVTSRPSYRPGGAPIQTHHGYNYRGRGRGRGTGGLRPVTKFTEDFDFMAMN 1546
Query: 513 EKFKKDEVWGHLGK----SNKKDGEENASDEDGGQDEDNGDVSNLEVKPVYNKDDFFDSL 568
EKFKKDEVWGHLGK S +KDGEENA DED QDEDN DVSN+EVK +YNKDDFFDSL
Sbjct: 1547EKFKKDEVWGHLGKSKSHSKEKDGEENAFDED-YQDEDNDDVSNIEVKLLYNKDDFFDSL 1723
Query: 569 SCNSLNHDPQNGRVRYSEQIKMDTETFGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGY 628
S N + QNGR RYSEQIK+DTETFGDF RH GG GRGP G GR RGGYYGR
Sbjct: 1724SSNMHGNASQNGRTRYSEQIKIDTETFGDFVRHHGGRRGRGP---GRVGRTRGGYYGR-L 1891
Query: 629 GGYGGRGRGGGRGM 642
GYG G G GRGM
Sbjct: 1892RGYGYNGGGRGRGM 1933
>TC226964 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (5%)
Length = 1717
Score = 543 bits (1400), Expect = e-155
Identities = 297/474 (62%), Positives = 333/474 (69%), Gaps = 7/474 (1%)
Frame = +3
Query: 177 PMYWQGYYGAPNGLPQLHQQSLFRPPPGLSMPSSMQQPMP-SSLQQPLPSSMQQPMPSSM 235
PMYWQGYYGAPNGLPQLHQQSL +PPPGLSMPSSMQQPM + PLP+ SS
Sbjct: 3 PMYWQGYYGAPNGLPQLHQQSLLQPPPGLSMPSSMQQPMQYPNFTPPLPT------VSSN 164
Query: 236 QQPMPSSMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPN 295
+PSS+ +P S P ++S P P S+LP PS P PSA
Sbjct: 165 LPELPSSL---LPVSASTPSVTSASLPPNLPPAPSALPPAPSALPPAPSA---------- 305
Query: 296 ITSTSAPPLNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSG 355
L P PSATLA E F SV+NKAP VS AA L +NLPSLA T+
Sbjct: 306 ---------------LSPVPSATLASEIFPVSVANKAPNVSTSAAMLASNLPSLALLTNP 440
Query: 356 GSDINAAV-PLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQ 414
DINA V P+++K NAISGSSL YQ+V Q++P++V SS S+ TE PSLVTP QLL+
Sbjct: 441 ARDINAIVPPISSKSNAISGSSLPYQSV-SQLSPAVVESSTSIHTETSAPSLVTPGQLLK 617
Query: 415 SGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVAAESQPPILPLPATSRPIHRPGGA 474
G + +S++PSQ PHKDVEVVQVSSTS+PEPSVPV+ E+QPPILPLP TSRP HRPGGA
Sbjct: 618 PGPIIVSSAQPSQAPHKDVEVVQVSSTSSPEPSVPVSVETQPPILPLPVTSRPNHRPGGA 797
Query: 475 SNQT-HHGYGYRGRGRGRGIGGFRPAERFTEDFDFTAMNEKFKKDEVWGHLGKSNK---- 529
QT HHGY YRGRGRGRG GGFR +FTEDFDFTAMNEKFKKDEVWGHLGKS
Sbjct: 798 PTQTHHHGYSYRGRGRGRGTGGFRSVTKFTEDFDFTAMNEKFKKDEVWGHLGKSKSHSKD 977
Query: 530 KDGEENASDEDGGQDEDNGDVSNLEVKPVYNKDDFFDSLSCNSLNHDPQNGRVRYSEQIK 589
+GEENA DED QDEDN DVSN+EVKPVYNKDDFFDSLS N + QNGR RYSEQIK
Sbjct: 978 NNGEENAFDED-YQDEDNDDVSNIEVKPVYNKDDFFDSLSSNMHGNASQNGRTRYSEQIK 1154
Query: 590 MDTETFGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMP 643
+DTETFGD+ R+ GG GGRGP G GGR RGGYYGRG GGYG GRG GRGMP
Sbjct: 1155IDTETFGDYVRYHGGRGGRGP---GRGGRTRGGYYGRG-GGYGYSGRGRGRGMP 1304
>TC217494 weakly similar to UP|Q8BSY2 (Q8BSY2) Mus musculus 11 days pregnant
adult female ovary and uterus cDNA, RIKEN full-length
enriched library, clone:5031405O05 product:metal
response element binding transcription factor 1, full
insert sequence, partial (3%)
Length = 1045
Score = 390 bits (1002), Expect = e-108
Identities = 212/338 (62%), Positives = 244/338 (71%), Gaps = 5/338 (1%)
Frame = +2
Query: 269 SSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQASV 328
S+SLP PS PSA P +S L+ +++ L PS TLA E SV
Sbjct: 8 SASLPPAPSALAPAPSALPPAPSS-----------LSSASSALSSVPSVTLASEILPVSV 154
Query: 329 SNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVP-LANKPNAISGSSLSYQTVPQQMT 387
+NKAP VS AA L +NL SL T G DINA VP ++ K +AISGSSL YQTV Q++
Sbjct: 155 TNKAPIVSTSAAMLASNLLSL---TISGPDINAIVPPISRKLHAISGSSLPYQTV-SQLS 322
Query: 388 PSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPS 447
++VGSS S+ TE PSL+TP QLLQ G ++ +S++PSQ PHK+VEVVQVSSTS+PEPS
Sbjct: 323 STVVGSSTSIHTETSAPSLLTPGQLLQPGPSIVSSAQPSQAPHKEVEVVQVSSTSSPEPS 502
Query: 448 VPVAAESQPPILPLPATSRPIHRPGGASNQTHHGYGYRGRGRGRGIGGFRPAERFTEDFD 507
VPV+AE+QPPILPLP TSRP +RPG A Q HHGY YRGRGRGRG GG P +FTEDFD
Sbjct: 503 VPVSAETQPPILPLPVTSRPSYRPGVAPIQIHHGYNYRGRGRGRGTGGLHPVTKFTEDFD 682
Query: 508 FTAMNEKFKKDEVWGHLGK----SNKKDGEENASDEDGGQDEDNGDVSNLEVKPVYNKDD 563
FTAMNEKFKKDEVWGHLGK S +KDGEENA DED QDEDN DVSN EVKP+YNKDD
Sbjct: 683 FTAMNEKFKKDEVWGHLGKSKSHSKEKDGEENAFDED-YQDEDNDDVSNFEVKPIYNKDD 859
Query: 564 FFDSLSCNSLNHDPQNGRVRYSEQIKMDTETFGDFSRH 601
FFDSLS N + QNGR RYSEQIK DTETFGDF +H
Sbjct: 860 FFDSLSSNVHGNTSQNGRTRYSEQIKNDTETFGDFVKH 973
Score = 37.7 bits (86), Expect = 0.016
Identities = 50/194 (25%), Positives = 72/194 (36%), Gaps = 25/194 (12%)
Frame = +2
Query: 197 SLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMP-SSMQQPM 255
SL P L+ S P PSSL + P + + +P S+ P S M
Sbjct: 14 SLPPAPSALAPAPSALPPAPSSLSSASSALSSVPSVTLASEILPVSVTNKAPIVSTSAAM 193
Query: 256 LSS--MQQPLQFPSFSSSLPTGPSNFPEFPSAFLP-----------VGTSSPNITSTSAP 302
L+S + + P ++ +P + LP VG+S+ T TSAP
Sbjct: 194 LASNLLSLTISGPDINAIVPPISRKLHAISGSSLPYQTVSQLSSTVVGSSTSIHTETSAP 373
Query: 303 PLNLSTTILPPAPSATLAPETFQA-----------SVSNKAPTVSLPAATLGANLPSLAP 351
L +L P PS + + QA S S+ P+V + A T LP P
Sbjct: 374 SLLTPGQLLQPGPSIVSSAQPSQAPHKEVEVVQVSSTSSPEPSVPVSAETQPPILP--LP 547
Query: 352 FTSGGSDINAAVPL 365
TS S P+
Sbjct: 548 VTSRPSYRPGVAPI 589
>TC226965 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, partial (21%)
Length = 973
Score = 276 bits (705), Expect = 3e-74
Identities = 141/191 (73%), Positives = 149/191 (77%), Gaps = 5/191 (2%)
Frame = +1
Query: 458 ILPLPATSRPIHRPGGASNQTHHGYGYRGRGRGRGIGGFRPAERFTEDFDFTAMNEKFKK 517
ILPLP TSRP +RPGGA +QTHHGY YRGRGRGRG GG P +FTEDFDF AMNEKFKK
Sbjct: 1 ILPLPVTSRPSYRPGGAPSQTHHGYNYRGRGRGRGTGGLHPVTKFTEDFDFMAMNEKFKK 180
Query: 518 DEVWGHLGK----SNKKDGEENASDEDGGQDEDNGDVSNLEVK-PVYNKDDFFDSLSCNS 572
DEVWGHLGK S +KDGEENA DED QDEDN DVSN+EVK P+YNKDDFFDSLS N
Sbjct: 181 DEVWGHLGKSKSHSKEKDGEENAFDED-YQDEDNDDVSNIEVKQPIYNKDDFFDSLSSNV 357
Query: 573 LNHDPQNGRVRYSEQIKMDTETFGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYG 632
+ QNGR RYSEQIK+DTETFGDF RHRGG RGG GGR RGGYYGR GGYG
Sbjct: 358 HGNASQNGRTRYSEQIKIDTETFGDFVRHRGG-------RGGRGGRTRGGYYGR--GGYG 510
Query: 633 GRGRGGGRGMP 643
GRG GRGMP
Sbjct: 511 YNGRGRGRGMP 543
>TC226966 similar to UP|Q9ASU0 (Q9ASU0) At1g26110/F28B23_21, partial (7%)
Length = 646
Score = 112 bits (280), Expect = 5e-25
Identities = 58/78 (74%), Positives = 59/78 (75%)
Frame = +1
Query: 566 DSLSCNSLNHDPQNGRVRYSEQIKMDTETFGDFSRHRGGWGGRGPWRGGGGGRARGGYYG 625
DSLS N QNGR RYSEQIK+DTETFGDF RHRGG GGRGP R GGR RGGYYG
Sbjct: 1 DSLSSNVHGXTSQNGRTRYSEQIKIDTETFGDFVRHRGGRGGRGPAR---GGRTRGGYYG 171
Query: 626 RGYGGYGGRGRGGGRGMP 643
RG GGYG GRG GRGMP
Sbjct: 172 RG-GGYGYNGRGRGRGMP 222
Score = 30.0 bits (66), Expect = 3.3
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +1
Query: 481 GYGYRGRGRGRGI 493
GYGY GRGRGRG+
Sbjct: 181 GYGYNGRGRGRGM 219
>TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, complete
Length = 1145
Score = 57.0 bits (136), Expect = 3e-08
Identities = 28/46 (60%), Positives = 32/46 (68%), Gaps = 1/46 (2%)
Frame = -3
Query: 596 GDFSRHRGGWGGRGPWRGGGGG-RARGGYYGRGYGGYGGRGRGGGR 640
G ++R GG+GGR GGGGG R+R G YG GYGG GG G GGGR
Sbjct: 366 GGYNRGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYGGGR 229
Score = 43.9 bits (102), Expect = 2e-04
Identities = 21/30 (70%), Positives = 21/30 (70%)
Frame = -3
Query: 612 RGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
RGGGGG GGY RG GGYGGR GGG G
Sbjct: 393 RGGGGGGGGGGY-NRGGGGYGGRSGGGGGG 307
Score = 43.1 bits (100), Expect = 4e-04
Identities = 29/61 (47%), Positives = 31/61 (50%), Gaps = 13/61 (21%)
Frame = -3
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYGR-----GYGGY--------GGRGRGGGRGMPGG 645
++ RGG GG G GGG R GGY GR G GGY GG G GGG G GG
Sbjct: 402 AQSRGGGGGGG---GGGYNRGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYGGG 232
Query: 646 R 646
R
Sbjct: 231 R 229
Score = 36.2 bits (82), Expect = 0.047
Identities = 22/55 (40%), Positives = 24/55 (43%), Gaps = 10/55 (18%)
Frame = -3
Query: 596 GDFSRHRGGWGGRGPWR----------GGGGGRARGGYYGRGYGGYGGRGRGGGR 640
G + GG GG G +R GGGGG GG RGY G G GG R
Sbjct: 345 GGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYGGGRDRGYSRGGDGGDGGWR 181
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 57.0 bits (136), Expect = 3e-08
Identities = 28/46 (60%), Positives = 32/46 (68%), Gaps = 1/46 (2%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWRGGGGG-RARGGYYGRGYGGYGGRGRGGGR 640
G ++R GG+GGR GGGGG R+R G YG GYGG GG G GGGR
Sbjct: 1511 GGYNRGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYGGGR 1648
Score = 45.4 bits (106), Expect = 8e-05
Identities = 20/29 (68%), Positives = 20/29 (68%)
Frame = +2
Query: 613 GGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
GGGGG GG Y RG GGYGGR GGG G
Sbjct: 1484 GGGGGGGGGGGYNRGGGGYGGRSGGGGGG 1570
Score = 43.9 bits (102), Expect = 2e-04
Identities = 31/63 (49%), Positives = 33/63 (52%), Gaps = 15/63 (23%)
Frame = +2
Query: 599 SRHRGGWGGRGPWRGGGGG--RARGGYYGR-----GYGGY--------GGRGRGGGRGMP 643
++ RGG GG G GGGGG R GGY GR G GGY GG G GGG G
Sbjct: 1469 AQSRGGGGGGG---GGGGGYNRGGGGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYG 1639
Query: 644 GGR 646
GGR
Sbjct: 1640 GGR 1648
Score = 36.2 bits (82), Expect = 0.047
Identities = 22/55 (40%), Positives = 24/55 (43%), Gaps = 10/55 (18%)
Frame = +2
Query: 596 GDFSRHRGGWGGRGPWR----------GGGGGRARGGYYGRGYGGYGGRGRGGGR 640
G + GG GG G +R GGGGG GG RGY G G GG R
Sbjct: 1532 GGYGGRSGGGGGGGGYRSRDGGYGGGYGGGGGGGYGGGRDRGYSRGGDGGDGGWR 1696
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 55.8 bits (133), Expect = 6e-08
Identities = 53/186 (28%), Positives = 79/186 (41%), Gaps = 4/186 (2%)
Frame = +1
Query: 202 PPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQ 261
PP P + +P+ + P PS +P+ + P PSS P+ + P
Sbjct: 31 PPIAPSPGNHPPYVPTPPKTPSPSYSPPNVPTPPKTPSPSSQPPFTPTPPKTP------S 192
Query: 262 PLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPN----ITSTSAPPLNLSTTILPPAPSA 317
P P + +PT PS + PS P T SP ST P +LS + PP+PS
Sbjct: 193 PTSQPPY---IPTPPSPISQPPSIATPPNTLSPTSQPPYPSTIPPSTSLSPSY-PPSPSP 360
Query: 318 TLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSL 377
+ AP T+ S AP S P+ + A P+++P + S P++ S S
Sbjct: 361 SSAP-TYPPSY--LAPATSPPSPSTSATTPTISPAATTPS-----------PSSSSSSGA 498
Query: 378 SYQTVP 383
S +T P
Sbjct: 499 SNETTP 516
Score = 33.9 bits (76), Expect = 0.23
Identities = 49/161 (30%), Positives = 59/161 (36%), Gaps = 5/161 (3%)
Frame = +1
Query: 312 PPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNA 371
P PS AP S N P V P T PS +P N P P
Sbjct: 4 PKTPSPIYAPP-IAPSPGNHPPYVPTPPKTPS---PSYSP-------PNVPTP----PKT 138
Query: 372 ISGSSLSYQTVPQQMTPSIVGSSNSVRTEP-PV---PSLVTPAQLLQ-SGQTVAASSKPS 426
S SS T TPS + T P P+ PS+ TP L + Q S+ P
Sbjct: 139 PSPSSQPPFTPTPPKTPSPTSQPPYIPTPPSPISQPPSIATPPNTLSPTSQPPYPSTIPP 318
Query: 427 QTPHKDVEVVQVSSTSAPEPSVPVAAESQPPILPLPATSRP 467
T +S + P PS P +A + PP PATS P
Sbjct: 319 ST--------SLSPSYPPSPS-PSSAPTYPPSYLAPATSPP 414
>TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(91%)
Length = 1885
Score = 55.8 bits (133), Expect = 6e-08
Identities = 61/264 (23%), Positives = 96/264 (36%), Gaps = 15/264 (5%)
Frame = +1
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLP-----SSMQQPMPSSMQQPMPSSMQQPMPSSMQQPM 255
P P S P++ P P+S P P S P P + PSS P+ SS
Sbjct: 226 PSPQTSPPTATSTPTPTSSPPPPPSPPGRSRSATPPPLAAPTASPSSSPAPLASSAATAP 405
Query: 256 LSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTS------APPLNLSTT 309
L S F ++S T P PV S N +S PP S++
Sbjct: 406 LPSKNAATAFSGSTTSTAT------TIPP*NAPVRQCSGNTRFSSWKVTLTTPPCLRSSS 567
Query: 310 ILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKP 369
L P+P+++ +P ++ + P + P L ++ S P P+ N P
Sbjct: 568 TLSPSPTSSTSPLRLASATPCRTPNPTSPPTLLASSTSSRLP----------NPPIPNPP 717
Query: 370 NAISGSSLSYQTVPQQMTPSIVGSSNSVRTEP----PVPSLVTPAQLLQSGQTVAASSKP 425
+ + S + P+ ++PS +N + P P TP + + ++S P
Sbjct: 718 *SGLHPAPSTASTPKTLSPSSTAPTNPPASTPPPRKPARRSPTPTTTSTASPSPDSASSP 897
Query: 426 SQTPHKDVEVVQVSSTSAPEPSVP 449
S P D + SS P
Sbjct: 898 STAPGGDPTWLTFSSPRTSSKGKP 969
Score = 53.1 bits (126), Expect = 4e-07
Identities = 58/211 (27%), Positives = 85/211 (39%), Gaps = 12/211 (5%)
Frame = +1
Query: 267 SFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQA 326
S SSS + PS P P TS+P TS+ PP + PP S + P
Sbjct: 184 SHSSSSSSSPSTTLPSPQTSPPTATSTPTPTSSPPPPPS------PPGRSRSATP----- 330
Query: 327 SVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSLS-YQTVPQQ 385
P ++ P A+ PS +P S A +P N A SGS+ S T+P
Sbjct: 331 ------PPLAAPTAS-----PSSSPAPLASSAATAPLPSKNAATAFSGSTTSTATTIPP* 477
Query: 386 MTPSIVGSSNS-------VRTEPPV---PSLVTPAQLLQSGQTVAASSKPSQTPH-KDVE 434
P S N+ T PP S ++P+ + AS+ P +TP+
Sbjct: 478 NAPVRQCSGNTRFSSWKVTLTTPPCLRSSSTLSPSPTSSTSPLRLASATPCRTPNPTSPP 657
Query: 435 VVQVSSTSAPEPSVPVAAESQPPILPLPATS 465
+ SSTS+ P+ P+ + P P+T+
Sbjct: 658 TLLASSTSSRLPNPPIPNPP*SGLHPAPSTA 750
Score = 33.9 bits (76), Expect = 0.23
Identities = 18/38 (47%), Positives = 18/38 (47%)
Frame = -2
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGR 640
G WGG G WR GG R R G G G GGGR
Sbjct: 354 GRWGGEG-WRSGGPASPRRRRRRRRRGSGSGSGGGGGR 244
Score = 29.6 bits (65), Expect = 4.4
Identities = 27/100 (27%), Positives = 46/100 (46%)
Frame = +1
Query: 348 SLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLV 407
S AP +S +++VPL+ ++ S SS S Q +P S+ + + PP P
Sbjct: 124 STAPNSSTPPPNSSSVPLS*SHSSSSSSSPSTTLPSPQTSPPTATSTPTPTSSPPPPP-- 297
Query: 408 TPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPS 447
+P +S T + P+ +P + S+ +AP PS
Sbjct: 298 SPPGRSRSA-TPPPLAAPTASPSSSPAPLASSAATAPLPS 414
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 55.1 bits (131), Expect = 1e-07
Identities = 59/211 (27%), Positives = 86/211 (39%), Gaps = 4/211 (1%)
Frame = +3
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLPSSMQ---QPMPSSMQQPMPSSMQQPMPSSMQQPMLS 257
P P +MP S P PS+ P S P PS P P S P ++ P S
Sbjct: 237 PSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSAS 416
Query: 258 SMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSA 317
P PS S T +N P S +SP+ TSTS PP S++ P PS+
Sbjct: 417 PPTSPS--PSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPSSSS--PA*PSS 584
Query: 318 TLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVP-LANKPNAISGSS 376
+P S + +P S P + S++P T+ S +P L + ++ S
Sbjct: 585 KPSPSPTPIS-PDPSPATSTPTSH-----TSISPITA--SKATFLLP*LCSTASSF*ISP 740
Query: 377 LSYQTVPQQMTPSIVGSSNSVRTEPPVPSLV 407
L+ + + + PS+ + PP PS V
Sbjct: 741 LT-PSPEKYLLPSVT*FHSKTSPSPPTPSPV 830
Score = 49.3 bits (116), Expect = 5e-06
Identities = 68/272 (25%), Positives = 99/272 (36%), Gaps = 19/272 (6%)
Frame = +3
Query: 197 SLFRPPPGLSMPSS-MQQPMPSSLQQPLP---SSMQQPMPSSMQQPMPSSMQQP---MPS 249
S F PP PS + + L P P + QQP+ S P + P MP
Sbjct: 84 SFFHPPLSNLSPSLILHLHLHLHLHHPHPLWTQNKQQPLNPSTSPPHRTPAPSPPSTMPP 263
Query: 250 SMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTT 309
S P S+ P S+ PT P P SP+ S+ +PP
Sbjct: 264 SATPPSPSATSSP-------SASPTAPPTSP------------SPSPPSSPSPP------ 368
Query: 310 ILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKP 369
P+PS+T + ++ +P+ PA+T AN P+ TS S + + +KP
Sbjct: 369 -SAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKP 545
Query: 370 NAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPV------PSLVTPAQLLQSGQTVAASS 423
A S SS + + +P+ + S T P P + A L ASS
Sbjct: 546 PAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISPITASKATFLLP*LCSTASS 725
Query: 424 ------KPSQTPHKDVEVVQVSSTSAPEPSVP 449
PS + V S ++P P P
Sbjct: 726 F*ISPLTPSPEKYLLPSVT*FHSKTSPSPPTP 821
Score = 39.3 bits (90), Expect = 0.006
Identities = 48/177 (27%), Positives = 64/177 (36%), Gaps = 26/177 (14%)
Frame = +3
Query: 186 APNGLPQLHQQSLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQ 245
+P+ P S PP P S P ++ P S P PS +S
Sbjct: 297 SPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPASTASANS 476
Query: 246 P---MPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPE-FPSAFLP---VGTSSPNITS 298
P P+S P S + P+ SSS P PS+ P P+ P TS+P +
Sbjct: 477 PASGSPTSRTSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPT-SH 653
Query: 299 TSAPPLNLS--TTILP---------------PAPSATLAPET--FQASVSNKAPTVS 336
TS P+ S T +LP P+P L P F + S PT S
Sbjct: 654 TSISPITASKATFLLP*LCSTASSF*ISPLTPSPEKYLLPSVT*FHSKTSPSPPTPS 824
Score = 32.3 bits (72), Expect = 0.67
Identities = 21/45 (46%), Positives = 22/45 (48%), Gaps = 6/45 (13%)
Frame = -1
Query: 607 GRGPWRGGGGGRARGGYYGRGYGGYGG------RGRGGGRGMPGG 645
GRG R GGG RG GRG GG+ G RGRG GG
Sbjct: 437 GRGRGRRGGGWG*RGS*GGRGSGGWRGT*GR*RRGRGRRSSWRGG 303
Score = 29.3 bits (64), Expect = 5.7
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 9/56 (16%)
Frame = -1
Query: 600 RHRGGWGGRGPWR-------GGGGGRAR--GGYYGRGYGGYGGRGRGGGRGMPGGR 646
R R G G G WR GG R R G GR GG+G RG GGRG G R
Sbjct: 527 RRR*GR*GSGGWRARRRRVCGGC*RR*RR*GRGRGRRGGGWG*RGS*GGRGSGGWR 360
Score = 28.5 bits (62), Expect = 9.7
Identities = 19/38 (50%), Positives = 20/38 (52%), Gaps = 7/38 (18%)
Frame = -1
Query: 596 GDFSRHRGGWGGRGPWRGG---GGGRA----RGGYYGR 626
G R R G G R WRGG GGGRA RG + GR
Sbjct: 359 GT*GR*RRGRGRRSSWRGGGR*GGGRAWRRRRGWHCGR 246
>TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1),
partial (3%)
Length = 784
Score = 55.1 bits (131), Expect = 1e-07
Identities = 56/197 (28%), Positives = 84/197 (42%), Gaps = 8/197 (4%)
Frame = +3
Query: 288 PVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLP 347
P SP+ T TS+ P N + +PSA+ T + S+ A T P +T P
Sbjct: 159 PSSPPSPSTTPTSSAPGNAPHS----SPSASKRRPT-PSGPSSAASTSPKPTSTSSRAAP 323
Query: 348 SLAPFT-----SGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPP 402
S +PFT G S + A P PNA + S+ S + + + S+ + + P
Sbjct: 324 SKSPFTWPSASLGTSMSSPASPPPPAPNASTSSTTSAASPALASSAANTASATTAPSLPS 503
Query: 403 VPSLVTP--AQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVAAESQPPILP 460
+PS TP A+ S +S+ P+ TP + + S+P PS + S PP
Sbjct: 504 IPSRTTPTTARSTPSSSNPTSSTSPTATPKR-------TRVSSPTPSSSSTSRSSPPSPK 662
Query: 461 LP-ATSRPIHRPGGASN 476
P AT H G +N
Sbjct: 663 EPTATVTVSHTRGEEAN 713
Score = 49.7 bits (117), Expect = 4e-06
Identities = 56/205 (27%), Positives = 87/205 (42%), Gaps = 2/205 (0%)
Frame = +3
Query: 202 PPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPS-SMQQPMPSSMQQPMLSSMQ 260
PP SS PS P S+ P + PS S ++P PS +S +
Sbjct: 129 PPASPRSSSPPSSPPSPSTTPTSSA-----PGNAPHSSPSASKRRPTPSGPSSAASTSPK 293
Query: 261 QPLQFPSFSSSLPTGPSNFP-EFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATL 319
S+S PS P +PSA L SSP ++PP PPAP+A+
Sbjct: 294 PT------STSSRAAPSKSPFTWPSASLGTSMSSP-----ASPP--------PPAPNAST 416
Query: 320 APETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSLSY 379
+ T AS + + + +AT +LPS+ P + + + +N ++ S ++
Sbjct: 417 SSTTSAASPALASSAANTASATTAPSLPSI-PSRTTPTTARSTPSSSNPTSSTSPTATPK 593
Query: 380 QTVPQQMTPSIVGSSNSVRTEPPVP 404
+T TPS SS++ R+ PP P
Sbjct: 594 RTRVSSPTPS---SSSTSRSSPPSP 659
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 54.3 bits (129), Expect = 2e-07
Identities = 63/211 (29%), Positives = 91/211 (42%), Gaps = 2/211 (0%)
Frame = +2
Query: 197 SLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPML 256
SL +PPP + P PM S P P+PS+ +++ P+S L
Sbjct: 176 SLSKPPP*PTTP-----PMLSPPGTP-------PLPSAPGSASRATLAAT*PAST*PRYL 319
Query: 257 SSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPS 316
S P+ P+F SS T PS P+ P+ SP S+++P +T PP+
Sbjct: 320 SPALYPMTSPTFRSS-RTSPSPTISSPAPSPPLSPPSPPSASSTSP----TTFSTPPSLP 484
Query: 317 ATLAPETFQASVSNKAPTVSLPAATLGANL--PSLAPFTSGGSDINAAVPLANKPNAISG 374
+++A T + S S T + P L +L PS A FTS + + P + P+ G
Sbjct: 485 SSIASPTLRFSTST---TTT*PENFLSPSLPCPSSATFTSAAT----SSPARSLPSTAPG 643
Query: 375 SSLSYQTVPQQMTPSIVGSSNSVRTEPPVPS 405
S+ S T P T S S S T PP S
Sbjct: 644 STSS--TSPSPATSSPAPSRPSSETSPPSAS 730
Score = 52.4 bits (124), Expect = 6e-07
Identities = 64/237 (27%), Positives = 93/237 (39%), Gaps = 11/237 (4%)
Frame = +2
Query: 242 SMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITST-- 299
S+ +P P PMLS P P PSA P S + +T
Sbjct: 176 SLSKPPP*PTTPPMLS------------------PPGTPPLPSA--PGSASRATLAAT*P 295
Query: 300 -SAPPLNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSD 358
S P LS + P + TF++S ++ +PT+S PA + + PS P S S
Sbjct: 296 AST*PRYLSPALYP------MTSPTFRSSRTSPSPTISSPAPSPPLSPPS-PPSASSTSP 454
Query: 359 INAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQT 418
+ P + P++I+ +L + T P N + P PS T + +
Sbjct: 455 TTFSTP-PSLPSSIASPTLRFSTSTTTT*PE-----NFLSPSLPCPSSAT---FTSAATS 607
Query: 419 VAASSKPSQTPHKDVEVVQVSSTSAPEPSVPVAAESQPP--------ILPLPATSRP 467
A S PS P +TS+P PS P ++E+ PP P PA SRP
Sbjct: 608 SPARSLPSTAPGSTSSTSPSPATSSPAPSRP-SSETSPPSASSTSATTTPTPAASRP 775
Score = 35.4 bits (80), Expect = 0.080
Identities = 20/47 (42%), Positives = 23/47 (48%), Gaps = 7/47 (14%)
Frame = -1
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYG-------GYGGRGRGGGRG 641
+G W + WRG G G RG RG+G G G RG GGRG
Sbjct: 481 KGRWR*KRCWRG*GSGGRRGRRKRRGWGRRTYCRRGRGARGTEGGRG 341
>TC203681 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 913
Score = 54.3 bits (129), Expect = 2e-07
Identities = 55/200 (27%), Positives = 82/200 (40%), Gaps = 7/200 (3%)
Frame = +3
Query: 289 VGTSSPNITSTSAPPLNLSTTILPPA--PSATLAPETFQASVSNKAPTVSLPAATLGANL 346
VG SP + T++PP +T PA PS +P + S+ P S AAT A
Sbjct: 66 VGGQSPAASPTTSPPAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAAT--ATP 239
Query: 347 PSLAP-FTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPS 405
P+ +P S S A P+A P A ++ P +TP + + PP P
Sbjct: 240 PASSPTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAPV 419
Query: 406 LVT--PAQLLQSGQTVAASSK--PSQTPHKDVEVVQVSSTSAPEPSVPVAAESQPPILPL 461
V+ PA + V A S P+ P + + +AP PS + PP+
Sbjct: 420 PVSSPPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKSKKHTAPAPSPSLLGPPAPPV-GA 596
Query: 462 PATSRPIHRPGGASNQTHHG 481
P +S+ PG A ++ G
Sbjct: 597 PGSSQDSMSPGPAVSEDESG 656
Score = 45.1 bits (105), Expect = 1e-04
Identities = 51/206 (24%), Positives = 75/206 (35%), Gaps = 5/206 (2%)
Frame = +3
Query: 228 QQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFL 287
Q P S P P++ P S P P+ P+ SS + P+ P A
Sbjct: 75 QSPAASPTTSP-PAATTTPFASPATAPSKPKSPAPVASPTSSSPPASSPNAATATPPASS 251
Query: 288 PVGTSSPNITSTSAPPLNLSTTILPPA--PSATLAPETFQASVSNKAPTVSLPAATLGAN 345
P S P+ + AP P A P+AT T +S P S PA ++
Sbjct: 252 PTVASPPSKAAAPAPVATPPAATPPAATPPAATPPAVTPVSSPPAPVPVSSPPAPVPVSS 431
Query: 346 LPSLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPS 405
P+LAP T +A A P + + S + +PS++G PP P
Sbjct: 432 PPALAPTTPAPVVAPSAEVPAPAPKSKKKTKKSKKHTAPAPSPSLLG--------PPAPP 587
Query: 406 LVTPAQLLQS---GQTVAASSKPSQT 428
+ P S G V+ ++T
Sbjct: 588 VGAPGSSQDSMSPGPAVSEDESGAET 665
Score = 43.1 bits (100), Expect = 4e-04
Identities = 38/151 (25%), Positives = 58/151 (38%)
Frame = +3
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQ 260
PP + P + PS + P P + P SS P++ P+S P ++S
Sbjct: 105 PPAATTTPFASPATAPSKPKSPAP--VASPTSSSPPASSPNAATATPPAS--SPTVASPP 272
Query: 261 QPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLA 320
P+ ++ P P A P + P +T S+PP + + PPAP +
Sbjct: 273 SKAAAPAPVATPPAAT------PPAATPPAATPPAVTPVSSPPAPVPVS-SPPAPVPVSS 431
Query: 321 PETFQASVSNKAPTVSLPAATLGANLPSLAP 351
P APT P A +P+ AP
Sbjct: 432 PPAL-------APTTPAPVVAPSAEVPAPAP 503
>TC203386 similar to UP|Q9XIV1 (Q9XIV1) Cucumis sativus mRNA expressed in
cucumber hypocotyls, complete cds (Arabinogalactan
protein), partial (40%)
Length = 649
Score = 53.5 bits (127), Expect = 3e-07
Identities = 60/228 (26%), Positives = 90/228 (39%), Gaps = 6/228 (2%)
Frame = +1
Query: 260 QQPLQFPSFSSSLPTGPSNFPEFPSAFLPVG--TSSPNITS----TSAPPLNLSTTILPP 313
Q P P+ SS+ P + P+ P + PV TSSP +S T+ PP++ T PP
Sbjct: 70 QSPAAAPTTSSASPATAPSKPK-PKSPAPVASPTSSPPASSPNAATATPPVSSPTVASPP 246
Query: 314 APSATLAPETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAIS 373
+ +A AP T P + PAAT A P P + S A VP+++ P +
Sbjct: 247 SKAAAPAPAT-------TPPVATPPAATPPAATP---PAVTPVSSPPAPVPVSSPPAPVP 396
Query: 374 GSSLSYQTVPQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDV 433
SS PP + TPA ++ V P+ P
Sbjct: 397 VSS------------------------PPALAPTTPAPVVAPSAEV-----PAPAPKSKK 489
Query: 434 EVVQVSSTSAPEPSVPVAAESQPPILPLPATSRPIHRPGGASNQTHHG 481
+ + +AP PS + PP+ P +S+ PG A ++ G
Sbjct: 490 KTKKSKKHTAPAPSPALLGPPAPPV-GAPGSSQDASSPGPAVSEDESG 630
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 52.4 bits (124), Expect = 6e-07
Identities = 30/51 (58%), Positives = 31/51 (59%), Gaps = 1/51 (1%)
Frame = +3
Query: 592 TETFGDFSR-HRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
T GD R + GG GG G GGGGG GG YG G GGYGG GRGGG G
Sbjct: 282 TRRGGDGGRSYGGGRGGGGGGYGGGGGYGGGGGYGGG-GGYGGGGRGGGGG 431
Score = 47.4 bits (111), Expect = 2e-05
Identities = 30/49 (61%), Positives = 30/49 (61%), Gaps = 5/49 (10%)
Frame = +3
Query: 602 RGGWGGR---GPWRGGGGGRARGGYYGRGYGGYGGRG--RGGGRGMPGG 645
RGG GGR G GGGGG GG YG G GGYGG G GGGRG GG
Sbjct: 288 RGGDGGRSYGGGRGGGGGGYGGGGGYGGG-GGYGGGGGYGGGGRGGGGG 431
Score = 43.5 bits (101), Expect = 3e-04
Identities = 22/38 (57%), Positives = 24/38 (62%)
Frame = +3
Query: 608 RGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
+G RGG GGR+ GG G G GGYGG G GG G GG
Sbjct: 276 QGTRRGGDGGRSYGGGRGGGGGGYGGGGGYGGGGGYGG 389
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/62 (45%), Positives = 30/62 (48%), Gaps = 17/62 (27%)
Frame = +3
Query: 603 GGWGGRGPW----RGGGGGR-------------ARGGYYGRGYGGYGGRGRGGGRGMPGG 645
GG+GG G + RGGGGG GG G YGG GG GR GG G GG
Sbjct: 375 GGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGGG--GG 548
Query: 646 RY 647
RY
Sbjct: 549 RY 554
Score = 39.3 bits (90), Expect = 0.006
Identities = 21/44 (47%), Positives = 23/44 (51%)
Frame = +3
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGG 639
G +R G GG + GGGGG GG G Y G GG G GGG
Sbjct: 456 GHLARDCSGGGGGDRYGGGGGGGRYGGGGGGRYVGGGGGGGGGG 587
Score = 37.7 bits (86), Expect = 0.016
Identities = 25/69 (36%), Positives = 27/69 (38%), Gaps = 26/69 (37%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGG--------------------------YGGRGR 636
GG+GG G + GGGG GGY G G GG YGG G
Sbjct: 339 GGYGGGGGYGGGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGG 518
Query: 637 GGGRGMPGG 645
GG G GG
Sbjct: 519 GGRYGGGGG 545
Score = 33.9 bits (76), Expect = 0.23
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Frame = +3
Query: 613 GGGGGRARGGYYGRGY-GGYGGRGRGGGRGMPGG 645
GGGG R GG G Y GG GGR GGG G GG
Sbjct: 483 GGGGDRYGGGGGGGRYGGGGGGRYVGGGGGGGGG 584
Score = 33.5 bits (75), Expect = 0.30
Identities = 20/40 (50%), Positives = 20/40 (50%), Gaps = 3/40 (7%)
Frame = +3
Query: 609 GPWRGGGGGRARGGYYGRGYG---GYGGRGRGGGRGMPGG 645
GP G RGG GR YG G GG G GGG G GG
Sbjct: 255 GPDGASVQGTRRGGDGGRSYGGGRGGGGGGYGGGGGYGGG 374
Score = 32.7 bits (73), Expect = 0.52
Identities = 27/65 (41%), Positives = 28/65 (42%), Gaps = 22/65 (33%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRAR-----------------GGYYGRGYGG--YGGRGRG---GGR 640
GG+GG G RGGGGG G YG G GG YGG G G GG
Sbjct: 393 GGYGGGG--RGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGGGGGRYVGGG 566
Query: 641 GMPGG 645
G GG
Sbjct: 567 GGGGG 581
Score = 30.0 bits (66), Expect = 3.3
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = +3
Query: 625 GRGYGGYGGRGRGGGRGMPGGRY 647
G GG GGR GGGRG GG Y
Sbjct: 279 GTRRGGDGGRSYGGGRGGGGGGY 347
>BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8). [Kidney bean French
bean], partial (11%)
Length = 169
Score = 52.4 bits (124), Expect = 6e-07
Identities = 26/50 (52%), Positives = 29/50 (58%)
Frame = -3
Query: 595 FGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPG 644
+GD H GG+GG G G GGG GG +G GYGG G G GGG G G
Sbjct: 164 YGDGGAHGGGYGG-GQGGGAGGGYGAGGEHGGGYGGGEGGGAGGGYGASG 18
Score = 35.8 bits (81), Expect = 0.061
Identities = 19/39 (48%), Positives = 20/39 (50%)
Frame = -3
Query: 607 GRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G G GGG G +GG G GYG G G G G G GG
Sbjct: 161 GDGGAHGGGYGGGQGGGAGGGYGAGGEHGGGYGGGEGGG 45
Score = 32.3 bits (72), Expect = 0.67
Identities = 15/28 (53%), Positives = 16/28 (56%)
Frame = -3
Query: 617 GRARGGYYGRGYGGYGGRGRGGGRGMPG 644
G GG +G GYGG G G GGG G G
Sbjct: 167 GYGDGGAHGGGYGGGQGGGAGGGYGAGG 84
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 52.0 bits (123), Expect = 8e-07
Identities = 30/52 (57%), Positives = 30/52 (57%), Gaps = 8/52 (15%)
Frame = +1
Query: 603 GGWGGRGPWRGGG---GGRARGGYYGR-----GYGGYGGRGRGGGRGMPGGR 646
GG GG G RGGG GGR GG Y R GYGG GG G GGG G GGR
Sbjct: 349 GGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGGGGGYGGGGGGGYGGGR 504
Score = 47.8 bits (112), Expect = 2e-05
Identities = 25/47 (53%), Positives = 27/47 (57%)
Frame = +1
Query: 599 SRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
++ RGG GG G GG GG GGY G G GG GG R GG G GG
Sbjct: 316 AQSRGGGGGGGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGG 456
Score = 44.7 bits (104), Expect = 1e-04
Identities = 28/53 (52%), Positives = 28/53 (52%), Gaps = 9/53 (16%)
Frame = +1
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGG---RGRGGG------RGMPGG 645
RGG GG GGGG GGY G G GGYGG RG GGG RG GG
Sbjct: 403 RGGGGGYNRSGGGGGYGGGGGYGGGGGGGYGGGRDRGYGGGGDRGYSRGGDGG 561
Score = 40.0 bits (92), Expect = 0.003
Identities = 28/56 (50%), Positives = 30/56 (53%), Gaps = 5/56 (8%)
Frame = +1
Query: 596 GDFSRHRGGWGGRGPWRGGGGGRARGGYYG---RGYGGYGGRG--RGGGRGMPGGR 646
G ++R GG G G GG GG GGY G RGYGG G RG RGG G G R
Sbjct: 415 GGYNRSGGGGGYGGG--GGYGGGGGGGYGGGRDRGYGGGGDRGYSRGGDGGDGGWR 576
Score = 32.0 bits (71), Expect = 0.88
Identities = 29/107 (27%), Positives = 37/107 (34%), Gaps = 5/107 (4%)
Frame = -3
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQ-PMLSSM 259
PPP P P P L P P P P P P P P + L+ M
Sbjct: 479 PPPPYPPPPP*PPPPPLRLYPPPPPRPPPPYPPPRPPPYPPPPPPPPPPPPRD*ASLTVM 300
Query: 260 QQPLQF----PSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAP 302
+P +F PS +S + + LPV S ST +P
Sbjct: 299 LRPSRFWPFIPSMASFMDCSEAKVTNPKPLDLPVSRSLIIFVSTISP 159
>TC228708 similar to UP|GRP2_PHAVU (P10496) Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8), partial (15%)
Length = 494
Score = 51.6 bits (122), Expect = 1e-06
Identities = 25/49 (51%), Positives = 30/49 (61%)
Frame = +3
Query: 595 FGDFSRHRGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMP 643
+GD H GG+GG G GGGGG GG +G G+GG GG G G G +P
Sbjct: 75 YGDGGAHGGGYGG-GAGAGGGGGYGAGGAHGGGFGGGGGSGGGHGGYVP 218
Score = 46.6 bits (109), Expect = 3e-05
Identities = 24/45 (53%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Frame = +3
Query: 601 HRGGWGGRGPWRGG-GGGRARGGYYGRGYGGYGGRGRGGGRGMPG 644
H G+GG G GG GGG GG +G GYGG G G GGG G G
Sbjct: 21 HGXGYGGAGGSGGGSGGGYGDGGAHGGGYGGGAGAGGGGGYGAGG 155
Score = 37.4 bits (85), Expect = 0.021
Identities = 28/65 (43%), Positives = 29/65 (44%), Gaps = 12/65 (18%)
Frame = +3
Query: 593 ETFGDFSRHRGGWGGRGPWRGGG---GGRARGGYYGR-------GYGGYG--GRGRGGGR 640
E G+ GG GG G GGG GG GGY G GYG G G G GGG
Sbjct: 6 EAGGEHGXGYGGAGGSGGGSGGGYGDGGAHGGGYGGGAGAGGGGGYGAGGAHGGGFGGGG 185
Query: 641 GMPGG 645
G GG
Sbjct: 186 GSGGG 200
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 51.6 bits (122), Expect = 1e-06
Identities = 52/162 (32%), Positives = 68/162 (41%), Gaps = 1/162 (0%)
Frame = +3
Query: 201 PPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQ 260
PPP S P + P S Q P P+S P PS + P PSS P PSS
Sbjct: 315 PPPVQSSPPPVPSSPPPS-QSPPPAST--PPPSPLSSPPPSS---PPPSSPPPSSPPPAS 476
Query: 261 QPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSA-PPLNLSTTILPPAPSATL 319
P P +S P+ P P P F P + P T A PP +L+ T+ P +
Sbjct: 477 PPPSSPPPASPPPSSPP--PSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVS 650
Query: 320 APETFQASVSNKAPTVSLPAATLGANLPSLAPFTSGGSDINA 361
+P + V AP +S P+ G PSL+ + G D A
Sbjct: 651 SPAPSPSKVF--APALS-PSLAPG---PSLSTISPSGDDSGA 758
Score = 48.9 bits (115), Expect = 7e-06
Identities = 46/148 (31%), Positives = 63/148 (42%), Gaps = 4/148 (2%)
Frame = +3
Query: 186 APNGLPQLHQQSLFRPPPGLSMPSSMQQPMPSSLQQPLPSSMQQPMPSSMQQPMPSSMQQ 245
+P +P S PP PS + P PSS P PSS P PSS P P+S
Sbjct: 333 SPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSS---PPPSS---PPPSS---PPPASPPP 485
Query: 246 PMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGTSSPNITSTSAPPLN 305
P P S P P FS T P P P +P + +P +T S+PP++
Sbjct: 486 SSPPPASPP--PSSPPPSSPPPFSPPPATPP---PATPPPAVPPPSLTPTVTPLSSPPVS 650
Query: 306 ----LSTTILPPAPSATLAPETFQASVS 329
+ + PA S +LAP +++S
Sbjct: 651 SPAPSPSKVFAPALSPSLAPGPSLSTIS 734
Score = 43.9 bits (102), Expect = 2e-04
Identities = 46/158 (29%), Positives = 62/158 (39%), Gaps = 8/158 (5%)
Frame = +3
Query: 222 PLPSSMQQPMPSSMQQPMPSSMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGP---SN 278
P P + P+ SS P+PSS P PS P + PL P SS P+ P S
Sbjct: 297 PTPQASPPPVQSS-PPPVPSS---PPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSP 464
Query: 279 FPEFPSAFLPVGTSSP--NITSTSAPPLNLSTTILPPA--PSATLAPETFQASVSNKAPT 334
P P P S P + +S PP + PPA P A P +P
Sbjct: 465 PPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPP 644
Query: 335 VSLPAATLG-ANLPSLAPFTSGGSDINAAVPLANKPNA 371
VS PA + P+L+P + G ++ P + A
Sbjct: 645 VSSPAPSPSKVFAPALSPSLAPGPSLSTISPSGDDSGA 758
Score = 35.0 bits (79), Expect = 0.10
Identities = 38/158 (24%), Positives = 56/158 (35%), Gaps = 11/158 (6%)
Frame = +3
Query: 232 PSSMQQPMPSSMQQPMPSSMQQPMLSSMQQPLQFPSFSSSLPTGPSNFPEFPSAFLPVGT 291
PS+ P P+ P P P + S P Q P +S+ P P + P P +
Sbjct: 276 PSTSPAP-PTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSP-------PPSS 431
Query: 292 SSPNITSTSA------PPLNLSTTILPPAPSATLAPETFQASVSNKAPTVSLPAATLGAN 345
P+ S+ PP + PP+ +P F + P PA +
Sbjct: 432 PPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSL 611
Query: 346 LPSLAPFTSGGSDINAAVPL-----ANKPNAISGSSLS 378
P++ P +S A P A P+ G SLS
Sbjct: 612 TPTVTPLSSPPVSSPAPSPSKVFAPALSPSLAPGPSLS 725
Score = 34.7 bits (78), Expect = 0.14
Identities = 36/136 (26%), Positives = 53/136 (38%), Gaps = 3/136 (2%)
Frame = +3
Query: 275 GPSNFPE--FPSAFLPVGTSSPNITSTSAPPLNLSTTILPPAPSATLAPETFQASVSNKA 332
GPS P P A P SSP +S PP P PS +P S+
Sbjct: 273 GPSTSPAPPTPQASPPPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPP 452
Query: 333 PTVSLPAATLGANLPSLAPFTSGGSDINAAVPLANKPNAISGSSLSYQTVPQQMTPSIVG 392
P+ PA+ ++ P +P S ++ P + P ++ P +TP++
Sbjct: 453 PSSPPPASPPPSSPPPASPPPSSPPP-SSPPPFSPPPATPPPATPPPAVPPPSLTPTVTP 629
Query: 393 -SSNSVRTEPPVPSLV 407
SS V + P PS V
Sbjct: 630 LSSPPVSSPAPSPSKV 677
Score = 33.1 bits (74), Expect = 0.40
Identities = 28/92 (30%), Positives = 34/92 (36%), Gaps = 3/92 (3%)
Frame = +3
Query: 383 PQQMTPSIVGSSNSVRTEPPVPSLVTPAQLLQSGQTVAASSKPSQTPHKDVEVVQVSSTS 442
P Q +P V SS PP S P+ L +S PS P +S
Sbjct: 321 PVQSSPPPVPSSPPPSQSPPPASTPPPSPL---SSPPPSSPPPSSPPPSSPPPASPPPSS 491
Query: 443 APEPSVPVAA---ESQPPILPLPATSRPIHRP 471
P S P ++ S PP P PAT P P
Sbjct: 492 PPPASPPPSSPPPSSPPPFSPPPATPPPATPP 587
Score = 31.6 bits (70), Expect = 1.2
Identities = 18/36 (50%), Positives = 20/36 (55%)
Frame = -1
Query: 605 WGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGR 640
WG G W GG R+ GG GR + GRGRG GR
Sbjct: 617 WGQ*GWWNCGG--RSGGGRSGRRW*KRRGRGRGRGR 516
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 50.4 bits (119), Expect = 2e-06
Identities = 26/43 (60%), Positives = 27/43 (62%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGG 645
G GGRG + GGGGR G YG G GGYGG G GGG G GG
Sbjct: 87 GXGGGRGGY--GGGGRGGRGGYGGGGGGYGGGGYGGGGGYGGG 209
Score = 50.1 bits (118), Expect = 3e-06
Identities = 27/46 (58%), Positives = 28/46 (60%)
Frame = +3
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGRY 647
RGG+GG G GG G GGY G GYGG GG G GGG G GG Y
Sbjct: 102 RGGYGGGGRGGRGGYGGGGGGYGGGGYGGGGGYGGGGGGG-GGGCY 236
Score = 41.2 bits (95), Expect = 0.001
Identities = 28/65 (43%), Positives = 29/65 (44%), Gaps = 22/65 (33%)
Frame = +3
Query: 603 GGWGGRGPWRGGGGGRARGGYYGRG-YGGYGGRGRGG---------------------GR 640
GG GGRG + GGGGG GGY G G YGG GG G GG GR
Sbjct: 120 GGRGGRGGYGGGGGGYGGGGYGGGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGGGGGR 299
Query: 641 GMPGG 645
GG
Sbjct: 300 YGGGG 314
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/40 (52%), Positives = 23/40 (57%)
Frame = +3
Query: 602 RGGWGGRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRG 641
RGG+GG G GGGG GGY G G GG GG + G G
Sbjct: 135 RGGYGGGGGGYGGGGYGGGGGYGGGGGGGGGGCYKCGETG 254
Score = 39.7 bits (91), Expect = 0.004
Identities = 27/53 (50%), Positives = 28/53 (51%), Gaps = 4/53 (7%)
Frame = +3
Query: 597 DFSRHRGGWGGRGPWRGG-GGGRARGGYYGRGYGG---YGGRGRGGGRGMPGG 645
D S +G GG G RG G G RGGY G G GG YGG G G G G GG
Sbjct: 30 DESPVQGTRGGGGXXRGSYGXGGGRGGYGGGGRGGRGGYGGGGGGYGGGGYGG 188
Score = 38.1 bits (87), Expect = 0.012
Identities = 27/52 (51%), Positives = 27/52 (51%), Gaps = 6/52 (11%)
Frame = +3
Query: 602 RGGWG------GRGPWRGGGGGRARGGYYGRGYGGYGGRGRGGGRGMPGGRY 647
RGG G G G RGG GG RGG GYGG GG G GGG GG Y
Sbjct: 54 RGGGGXXRGSYGXGGGRGGYGGGGRGGR--GGYGG-GGGGYGGGGYGGGGGY 200
Score = 35.4 bits (80), Expect = 0.080
Identities = 25/63 (39%), Positives = 28/63 (43%), Gaps = 13/63 (20%)
Frame = +3
Query: 596 GDFSRHRGGWGGR----GPWRGGGGGRARGGYYGRGYGGY---------GGRGRGGGRGM 642
G + GG+GG G GGGGG GG Y G G+ GG GR GG G
Sbjct: 138 GGYGGGGGGYGGGGYGGGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGGGGGRYGGGGG 317
Query: 643 PGG 645
GG
Sbjct: 318 GGG 326
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.130 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,051,110
Number of Sequences: 63676
Number of extensions: 550918
Number of successful extensions: 12840
Number of sequences better than 10.0: 1093
Number of HSP's better than 10.0 without gapping: 6739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10272
length of query: 647
length of database: 12,639,632
effective HSP length: 103
effective length of query: 544
effective length of database: 6,081,004
effective search space: 3308066176
effective search space used: 3308066176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146570.8


