

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.7 - phase: 0
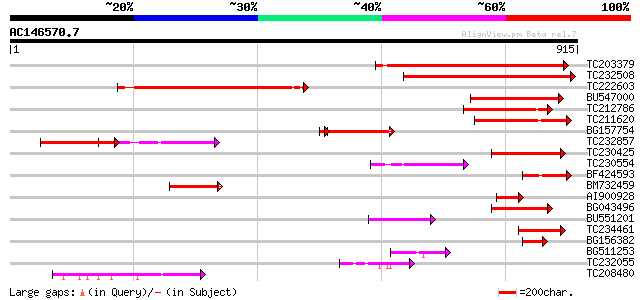
(915 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 479 e-135
TC232508 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 441 e-124
TC222603 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 392 e-109
BU547000 similar to GP|29467527|db P0582D05.17 {Oryza sativa (ja... 189 4e-48
TC212786 similar to UP|O49524 (O49524) Pherophorin - like protei... 166 3e-41
TC211620 similar to UP|O49524 (O49524) Pherophorin - like protei... 163 3e-40
BG157754 132 2e-33
TC232857 similar to UP|Q96258 (Q96258) AR791, partial (14%) 120 3e-27
TC230425 weakly similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_2... 112 7e-25
TC230554 similar to UP|Q9SX62 (Q9SX62) F11A17.16, partial (24%) 101 2e-21
BF424593 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arab... 87 4e-17
BM732459 similar to GP|1669599|dbj| AR791 {Arabidopsis thaliana}... 79 1e-14
AI900928 homologue to GP|28071265|db actin binding protein {Arab... 78 2e-14
BG043496 weakly similar to GP|15983793|gb| At1g48280/F11A17_25 {... 75 1e-13
BU551201 similar to GP|3261822|emb| PE_PGRS {Mycobacterium tuber... 74 3e-13
TC234461 similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_25, part... 73 6e-13
BG156382 similar to GP|28071265|db actin binding protein {Arabid... 69 9e-12
BG511253 similar to PIR|G96522|G965 F11A17.16 [imported] - Arabi... 65 2e-10
TC232055 similar to UP|O49524 (O49524) Pherophorin - like protei... 60 6e-09
TC208480 53 5e-07
>TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (29%)
Length = 1246
Score = 479 bits (1234), Expect = e-135
Identities = 247/315 (78%), Positives = 268/315 (84%), Gaps = 3/315 (0%)
Frame = +2
Query: 591 PPPKPSDGAPVSNSLNEIPYAPSVPSPPPPPGSLPRGAVGDDKVKRAPELVEFYQSLMKR 650
PPP P GAP P P PPPPPGSL RG + DKV RAP+LVEFYQ+LMKR
Sbjct: 44 PPPPPPPGAPPPP-----PPPGGPPPPPPPPGSLSRGGMDGDKVHRAPQLVEFYQTLMKR 208
Query: 651 EAKKD--ASLLTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQGDFVMSLATEVRAA 708
EAKKD +SLL +S SNA+D RSN+I EIENRSSFLLAVKADVETQGDFVMSLA EVRAA
Sbjct: 209 EAKKDTSSSLLVTSASNASDARSNMIGEIENRSSFLLAVKADVETQGDFVMSLAAEVRAA 388
Query: 709 SFSKIEDVVAFVNWLDEELSFLSDERAVLKHFDWPEGKSDALREASFEYQDLMKLEKQVS 768
SFS I D+VAFVNWLDEELSFL DERAVLKHFDWPEGK+DALREA+FEYQDLMKLE +VS
Sbjct: 389 SFSDINDLVAFVNWLDEELSFLVDERAVLKHFDWPEGKADALREAAFEYQDLMKLENRVS 568
Query: 769 NFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGK 828
F DDP LPCE AL+KMYSLLEK+EQSVYALLRTRD AISRYKEFG+PVNWL+DSGVVGK
Sbjct: 569 TFVDDPNLPCEAALKKMYSLLEKVEQSVYALLRTRDMAISRYKEFGIPVNWLMDSGVVGK 748
Query: 829 IKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKA 888
IKLSSVQLA KYMKR+ASE+D LSGP+ EP REFL+LQGVRF+FRVHQFAGGFD ESMKA
Sbjct: 749 IKLSSVQLAKKYMKRVASELDELSGPDKEPAREFLVLQGVRFAFRVHQFAGGFDAESMKA 928
Query: 889 FEELRNNIHV-QAGE 902
FEELR+ I QAGE
Sbjct: 929 FEELRSRIQTSQAGE 973
>TC232508 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (25%)
Length = 1050
Score = 441 bits (1134), Expect = e-124
Identities = 228/281 (81%), Positives = 251/281 (89%), Gaps = 3/281 (1%)
Frame = +2
Query: 636 RAPELVEFYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQG 695
RAPELVEFYQ+LMK EAKKD SL+ SST+ D RSN+I EIENRSSFLLAVKADVETQG
Sbjct: 2 RAPELVEFYQTLMKLEAKKDTSLI-SSTTYPFDARSNMIGEIENRSSFLLAVKADVETQG 178
Query: 696 DFVMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHFDWPEGKSDALREASF 755
DFV+SLATEVRAASFSKIED+VAFVNWLDEELSFL DE+AVLKHFDWPEGK+DA+REA+F
Sbjct: 179 DFVISLATEVRAASFSKIEDLVAFVNWLDEELSFLVDEQAVLKHFDWPEGKTDAMREAAF 358
Query: 756 EYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKEFGV 815
EY DLMKLEKQVS FTDDPKLPC+ ALQKMYSLLEK+EQS+YALLRTRD AISRYKEFG+
Sbjct: 359 EYLDLMKLEKQVSTFTDDPKLPCQTALQKMYSLLEKVEQSIYALLRTRDMAISRYKEFGI 538
Query: 816 PVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVH 875
PV WLLD+G+VGKIKLSSVQLAN YMKR+ASE++ LSGP+ EPTREFLILQGV F+FRVH
Sbjct: 539 PVTWLLDTGLVGKIKLSSVQLANMYMKRVASELEILSGPKKEPTREFLILQGVHFAFRVH 718
Query: 876 QFAGGFDTESMKAFEELRNNIHV-QAGEYNN--KPEMATKD 913
QFAGGFDTESMK FEELR+ IH GE NN K +TKD
Sbjct: 719 QFAGGFDTESMKVFEELRSRIHAPPPGEDNNNQKRS*STKD 841
>TC222603 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (27%)
Length = 881
Score = 392 bits (1008), Expect = e-109
Identities = 215/309 (69%), Positives = 244/309 (78%), Gaps = 1/309 (0%)
Frame = +2
Query: 175 KEEEVVKKDAEIENNLKTVNDFEIEKKLKTVNDLEIEAVGLKRRNKELQHEKRELTVKLN 234
KEEE +KDAE+E KKLK VNDLE+ V LKR+NKELQHEKRELTVKLN
Sbjct: 2 KEEEAARKDAEVE------------KKLKAVNDLEVAVVELKRKNKELQHEKRELTVKLN 145
Query: 235 AAESRITELSSVTENEMIADAKSETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVN 294
AESR ELS++TE+EM+A AK E LRHANEDL KQVEGLQMNRFSEVEELVYLRWVN
Sbjct: 146 VAESRAAELSNMTESEMVAKAKEEVSNLRHANEDLLKQVEGLQMNRFSEVEELVYLRWVN 325
Query: 295 ACLRYELRNYKAPSGKSLARDLNNSFNPKSQEKAKQLMLEYAGSERGHGDTDIESNFSHD 354
ACLRYELRN + P GK ARDL+ S +PKSQEKAKQLMLEYAGSERG GDTD+ESNFSH
Sbjct: 326 ACLRYELRNNQTPQGKVSARDLSKSLSPKSQEKAKQLMLEYAGSERGQGDTDLESNFSHP 505
Query: 355 HSSPGSEDLDNAYINSPTYKYSNLSKKTSLIQKLKKWNKN-NDSSAFSSPARSLSIGSPC 413
SSPGSED DNA I+S T KYS+LSKKTSLIQK KKW K+ +DSSA SSPARS S GSP
Sbjct: 506 -SSPGSEDFDNASIDSSTSKYSSLSKKTSLIQKFKKWGKSKDDSSALSSPARSFSGGSPR 682
Query: 414 RVSTSYRPRNTLESLMLRNAHDAVAITTFEQKEHKPTHSPETPFLPSLRKVSSSDDILDS 473
R+S S + R LESLMLRNA D+V+IT+F ++ +PT SPETP +R+V SSD L+S
Sbjct: 683 RMSVSVKQRGPLESLMLRNASDSVSITSFGLRDQEPTDSPETP--NDMRRVPSSDS-LNS 853
Query: 474 VSASFQLMS 482
V++SFQLMS
Sbjct: 854 VASSFQLMS 880
>BU547000 similar to GP|29467527|db P0582D05.17 {Oryza sativa (japonica
cultivar-group)}, partial (19%)
Length = 695
Score = 189 bits (481), Expect = 4e-48
Identities = 89/151 (58%), Positives = 118/151 (77%)
Frame = -2
Query: 744 EGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTR 803
E K+D LREA+F YQDL KLE +V + DDP+LP + A +KM +L EK+E++VY LLRTR
Sbjct: 694 EKKADTLREAAFGYQDLKKLEXEVXXYKDDPRLPGDIAXKKMVALSEKMERTVYNLLRTR 515
Query: 804 DFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFL 863
D + KEF +P+ W+LD+G++GKIKLSSV+LA KYMKR+A E+ S E +P +++
Sbjct: 514 DSLMRHSKEFKIPIEWMLDNGIIGKIKLSSVKLAKKYMKRVAMELQVKSALEKDPAMDYM 335
Query: 864 ILQGVRFSFRVHQFAGGFDTESMKAFEELRN 894
+LQGVRF+FR+HQFAGGFD E+M AFEELRN
Sbjct: 334 LLQGVRFAFRIHQFAGGFDAETMHAFEELRN 242
>TC212786 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(22%)
Length = 430
Score = 166 bits (421), Expect = 3e-41
Identities = 81/144 (56%), Positives = 109/144 (75%)
Frame = +3
Query: 733 ERAVLKHFDWPEGKSDALREASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKL 792
ERAVLKHFDWPE K+DALREA+F Y DL KLE + S+F DDP+ PC AL+KM L EKL
Sbjct: 3 ERAVLKHFDWPEQKADALREAAFGYCDLKKLESEASSFRDDPRQPCGPALKKMQVLFEKL 182
Query: 793 EQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLS 852
E V+ + R R+ A +RYK F +PV+W+LD+G V ++KL+SV+LA KYM+R+++E++T
Sbjct: 183 EHGVFNISRMRESATNRYKVFHIPVHWMLDNGFVSQMKLASVKLAMKYMRRVSAELET-- 356
Query: 853 GPENEPTREFLILQGVRFSFRVHQ 876
G P E +++QGVRF+FR HQ
Sbjct: 357 GGGCGPEEEEIVVQGVRFAFRAHQ 428
>TC211620 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(24%)
Length = 676
Score = 163 bits (413), Expect = 3e-40
Identities = 86/159 (54%), Positives = 113/159 (70%), Gaps = 3/159 (1%)
Frame = +2
Query: 751 REASFEYQDLMKLEKQVSNFTDDPKLPCEDALQKMYSLLEKLEQSVYALLRTRDFAISRY 810
REA+F Y DL KL + S+F DDP+ C AL+KM +LLEKLE +Y + R R+ A RY
Sbjct: 2 REAAFGYCDLKKLVSEASSFRDDPRQLCGPALKKMQTLLEKLEHGIYNISRMRESATKRY 181
Query: 811 KEFGVPVNWLLDSGVVGKIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRF 870
K F +PV+W+LDSG V +IKL+SV+LA KYMKR+++E++T G P E LI+Q VRF
Sbjct: 182 KVFQIPVDWMLDSGYVSQIKLASVKLAMKYMKRVSAELETGGG---GPEEEELIVQVVRF 352
Query: 871 SFRVHQFAGGFDTESMKAFEELRN---NIHVQAGEYNNK 906
+FRVHQFAGGFD E+M+AF+ELR+ + HVQ K
Sbjct: 353 AFRVHQFAGGFDVETMRAFQELRDKARSCHVQCHSQQQK 469
>BG157754
Length = 370
Score = 132 bits (332), Expect(2) = 2e-33
Identities = 71/111 (63%), Positives = 82/111 (72%), Gaps = 3/111 (2%)
Frame = +1
Query: 513 KAEKARGEKFGDNSNLNTTKAEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDNQT 572
KAEKAR EKFGDNSNLN TKAE ER+TSL +LTQ+ EKA VS N ++VK+ DNQT
Sbjct: 43 KAEKARAEKFGDNSNLNMTKAERERTTSLLPKLTQLKEKAYVSGSPNDHPDNVKNVDNQT 222
Query: 573 ITEMKLFKIEKRPPRLPRPPPKPSDGAPV---SNSLNEIPYAPSVPSPPPP 620
I++MKL IEKRPP +P+PPP PSDGAPV SN N + AP P PPPP
Sbjct: 223 ISKMKLAHIEKRPPXVPQPPP*PSDGAPVSANSNPSNGVSCAP--PQPPPP 369
Score = 29.6 bits (65), Expect(2) = 2e-33
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = +2
Query: 500 HKLALAREKQLKEKAEK 516
HKLAL REKQ+KEK K
Sbjct: 5 HKLALQREKQIKEKLRK 55
>TC232857 similar to UP|Q96258 (Q96258) AR791, partial (14%)
Length = 892
Score = 120 bits (301), Expect = 3e-27
Identities = 73/195 (37%), Positives = 116/195 (59%)
Frame = +1
Query: 144 IKELQRQIKIIANQTKGQLLLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLK 203
I++L ++ II +++ LL +K G + K+ + V D +I+ ++ D
Sbjct: 304 IRKLGTKLNIIGSRS*----LLSRKFPGCKTKKCKDVACDPDIQITMQKXKD-------- 447
Query: 204 TVNDLEIEAVGLKRRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLR 263
LE EA L++ N LQ E +L +L++ + ++ E+ K E+ L+
Sbjct: 448 ----LESEAEELRKSNLRLQIENSDLARRLDSTQILA---NAFLEDPEAGAVKQESECLK 606
Query: 264 HANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLNNSFNPK 323
N L K++E Q +R S++EELVYLRW+NACLRYELRNY+AP GK++A+DL+ S +P
Sbjct: 607 QENVRLMKEIEQFQSDRCSDLEELVYLRWINACLRYELRNYQAPPGKTVAKDLSRSLSPM 786
Query: 324 SQEKAKQLMLEYAGS 338
S++KAKQL+LEYA +
Sbjct: 787 SEKKAKQLILEYANA 831
Score = 88.2 bits (217), Expect = 1e-17
Identities = 45/126 (35%), Positives = 80/126 (62%)
Frame = +3
Query: 51 KNGSDSEIEWLRNVVEELEEREMKLQSELLEYYSLKEQVPVIEEFQRQLRIKSVEIDMLH 110
K+ + EI LR+++ L++R L+ +LLEY L+EQ + E Q +L+ ++E+ + +
Sbjct: 12 KDDYEQEISQLRSMIRMLQDRGRSLEVQLLEYCRLREQETAVIELQNRLKASTMEVKIFN 191
Query: 111 MTIKSLQEENNKLQEELIHEASAKRELEVARNKIKELQRQIKIIANQTKGQLLLLKQKVS 170
+ +K+LQ EN +L+E++ A ELE A+ ++K L ++I+ A + Q++ LKQKVS
Sbjct: 192 LKVKTLQSENWRLKEQVAGHAKVLAELETAKAQVKLLNKKIRHEAEHNREQIITLKQKVS 371
Query: 171 GLQAKE 176
LQ +E
Sbjct: 372 RLQDQE 389
>TC230425 weakly similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_25, partial
(61%)
Length = 784
Score = 112 bits (280), Expect = 7e-25
Identities = 49/119 (41%), Positives = 80/119 (67%)
Frame = +1
Query: 778 CEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLA 837
C AL+KM SLL+K E+S+ L++ R Y+ + +P W+LDSG++ +IK +S+ L
Sbjct: 22 CGAALKKMASLLDKSERSIQRLIKLRSSVTHSYQMYNIPTAWMLDSGIMSEIKQASMTLV 201
Query: 838 NKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMKAFEELRNNI 896
YMKR+ E++++ + E ++ L+LQG+ F++R HQF GG D+E+M AFEE+R +
Sbjct: 202 KTYMKRVTMELESIRNSDRESIQDSLLLQGMHFAYRAHQFTGGLDSETMCAFEEIRQRV 378
>TC230554 similar to UP|Q9SX62 (Q9SX62) F11A17.16, partial (24%)
Length = 1106
Score = 101 bits (251), Expect = 2e-21
Identities = 62/163 (38%), Positives = 95/163 (58%), Gaps = 5/163 (3%)
Frame = +2
Query: 583 KRPPR--LPRPPPKPSDG--APVSNSLNEIPYAPSVPSPPPPPGSLPRGAVGDDKVKRAP 638
K PP L PPP S G +P + L P PPPP + P + + + ++P
Sbjct: 644 KSPPNTCLQPPPPVTSVGRKSPSNTCLQ--------PPPPPPIPTRPLARLANSQ--KSP 793
Query: 639 ELVEFYQSLMKREAKKDAS-LLTSSTSNAADTRSNVIAEIENRSSFLLAVKADVETQGDF 697
+VE + SL ++ K D+ + S+++ EI+NRS+ LLA++AD+ET+G+F
Sbjct: 794 AIVELFHSLKNKDWKIDSKGSVNHQRPVVISAHSSIVGEIQNRSAHLLAIRADIETKGEF 973
Query: 698 VMSLATEVRAASFSKIEDVVAFVNWLDEELSFLSDERAVLKHF 740
+ L +V A+F+ IE+V+ FV+WLD +LS L+DERAVLK F
Sbjct: 974 INDLIRKVVDAAFTDIEEVLKFVDWLDVKLSSLADERAVLKPF 1102
>BF424593 homologue to GP|22135814|gb| AT4g18560/F28J12_220 {Arabidopsis
thaliana}, partial (11%)
Length = 411
Score = 86.7 bits (213), Expect = 4e-17
Identities = 46/82 (56%), Positives = 61/82 (74%), Gaps = 3/82 (3%)
Frame = +3
Query: 828 KIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGGFDTESMK 887
+IKL+SV+LA KYMKR+++E++T G P E LI+QGVRF+FRVHQFAGGFD E+M+
Sbjct: 111 QIKLASVKLAMKYMKRVSAELETGGGG---PEEEELIVQGVRFAFRVHQFAGGFDVETMR 281
Query: 888 AFEELRN---NIHVQAGEYNNK 906
AF+ELR+ + HVQ K
Sbjct: 282 AFQELRDKARSCHVQCHSQQQK 347
>BM732459 similar to GP|1669599|dbj| AR791 {Arabidopsis thaliana}, partial
(12%)
Length = 435
Score = 78.6 bits (192), Expect = 1e-14
Identities = 41/86 (47%), Positives = 56/86 (64%)
Frame = +3
Query: 258 ETGRLRHANEDLQKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARDLN 317
E RLR NE L K++E L +R ++EELVYLRW+NACLR+ELR+Y+ P GK++ARDL+
Sbjct: 3 EGERLRRENEGLTKELEQLHADRCLDLEELVYLRWINACLRHELRSYQPPPGKTVARDLS 182
Query: 318 NSFNPKSQEKAKQLMLEYAGSERGHG 343
S + K L L + +G G
Sbjct: 183 KSLIFHLRRKPSNLYLNMQAT-KGEG 257
Score = 34.3 bits (77), Expect = 0.26
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 5/75 (6%)
Frame = +2
Query: 321 NPKSQEKAKQLMLEYAGSERGHGDTDIESN-FSHDHSS----PGSEDLDNAYINSPTYKY 375
N S++KAKQL+LEYA +E +D++S+ +S +S PG + NS K
Sbjct: 191 NFSSEKKAKQLILEYASNEGRGSVSDMDSDQWSSSQASFLTDPGEREDYFPLDNSSELKA 370
Query: 376 SNLSKKTSLIQKLKK 390
+N + K+ + KL +
Sbjct: 371 TNNTSKSRIFGKLMR 415
>AI900928 homologue to GP|28071265|db actin binding protein {Arabidopsis
thaliana}, partial (3%)
Length = 425
Score = 78.2 bits (191), Expect = 2e-14
Identities = 35/44 (79%), Positives = 40/44 (90%)
Frame = -2
Query: 786 YSLLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKI 829
Y L ++EQSVYALLRTRD AISRYKEFG+PVNWL+DSGVVGK+
Sbjct: 280 YVFLSRVEQSVYALLRTRDMAISRYKEFGIPVNWLMDSGVVGKV 149
>BG043496 weakly similar to GP|15983793|gb| At1g48280/F11A17_25 {Arabidopsis
thaliana}, partial (43%)
Length = 423
Score = 75.5 bits (184), Expect = 1e-13
Identities = 32/99 (32%), Positives = 60/99 (60%)
Frame = +1
Query: 778 CEDALQKMYSLLEKLEQSVYALLRTRDFAISRYKEFGVPVNWLLDSGVVGKIKLSSVQLA 837
C L + S ++ E+S+ L++ R Y+ + +P W+LDSG++ +IK +S+ L
Sbjct: 13 CMLTLSLISSHCDRSERSIQRLIKLRSSVTHSYQMYNIPTAWMLDSGIMSEIKQASMTLV 192
Query: 838 NKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQ 876
YMKR+ E++++ + E ++ L+LQG+ F++R HQ
Sbjct: 193 KTYMKRVTMELESIRNSDRESIQDSLLLQGMHFAYRAHQ 309
>BU551201 similar to GP|3261822|emb| PE_PGRS {Mycobacterium tuberculosis
H37Rv}, partial (7%)
Length = 689
Score = 73.9 bits (180), Expect = 3e-13
Identities = 43/110 (39%), Positives = 66/110 (59%), Gaps = 2/110 (1%)
Frame = +3
Query: 580 KIEKRPPRLPRPPPKPSDGAPVSNSLNEIPYAP-SVP-SPPPPPGSLPRGAVGDDKVKRA 637
K++ R ++P P P + P + + P ++P + PPPP + P+ VG V+R
Sbjct: 126 KVKDRSVKVPPPAPSSNPLLPSHKTEKGMKVQPLALPRTAPPPPPTPPKSLVGLKSVRRV 305
Query: 638 PELVEFYQSLMKREAKKDASLLTSSTSNAADTRSNVIAEIENRSSFLLAV 687
PE++E Y+SL +++A D + T+ T AA TR N+I EIENRS+FL AV
Sbjct: 306 PEVIELYRSLTRKDANNDNKISTNGTPAAAFTR-NMIEEIENRSTFLSAV 452
>TC234461 similar to UP|Q93ZL7 (Q93ZL7) At1g48280/F11A17_25, partial (20%)
Length = 412
Score = 72.8 bits (177), Expect = 6e-13
Identities = 34/76 (44%), Positives = 53/76 (69%)
Frame = +3
Query: 821 LDSGVVGKIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGVRFSFRVHQFAGG 880
LDSG++ KIK +S+ L YMKR+ E+ + + ++E L+LQG+ F++R HQFAGG
Sbjct: 3 LDSGIMTKIKQASMILVKMYMKRVTMELGSARNSDRS-SQESLLLQGMHFAYRAHQFAGG 179
Query: 881 FDTESMKAFEELRNNI 896
D E++ AFEE+R ++
Sbjct: 180 LDAETLCAFEEIRQHV 227
>BG156382 similar to GP|28071265|db actin binding protein {Arabidopsis
thaliana}, partial (3%)
Length = 369
Score = 68.9 bits (167), Expect = 9e-12
Identities = 33/41 (80%), Positives = 38/41 (92%)
Frame = +3
Query: 828 KIKLSSVQLANKYMKRIASEIDTLSGPENEPTREFLILQGV 868
+IKLSSVQLAN YMKR+ASE++ LSGP+ EPTREFLILQGV
Sbjct: 246 QIKLSSVQLANMYMKRVASELEILSGPKKEPTREFLILQGV 368
>BG511253 similar to PIR|G96522|G965 F11A17.16 [imported] - Arabidopsis
thaliana, partial (6%)
Length = 433
Score = 64.7 bits (156), Expect = 2e-10
Identities = 35/102 (34%), Positives = 59/102 (57%), Gaps = 5/102 (4%)
Frame = +2
Query: 615 PSPPPPPGSLPRGAVGD-DKVKRAPELVEFYQSLMKREAKKDASLLTSSTSN----AADT 669
P PPP P S+P + + +RAP V+ + +L +E K T S A +
Sbjct: 134 PPPPPMPPSIPSRPIAKANNTQRAPAFVKLFHTLKNQEGMKST---TGSGKQQRPVAVNV 304
Query: 670 RSNVIAEIENRSSFLLAVKADVETQGDFVMSLATEVRAASFS 711
S+++ EI+NRS+ LLA++AD+ET+G+F+ L +V A+++
Sbjct: 305 HSSIVGEIQNRSAHLLAIRADIETKGEFINDLIKKVVEAAYT 430
>TC232055 similar to UP|O49524 (O49524) Pherophorin - like protein, partial
(7%)
Length = 448
Score = 59.7 bits (143), Expect = 6e-09
Identities = 45/140 (32%), Positives = 66/140 (47%), Gaps = 20/140 (14%)
Frame = +2
Query: 533 AEEERSTSLDSELTQVNEKACVSDGLNKQSEDVKDFDNQTITEMKLFKIEKRPPRLPRPP 592
A + S +L E+++++E+ S SE++ D + + + R PR+P PP
Sbjct: 5 ASDLGSGNLKKEVSEISERPRHS---RCNSEELVDCSDSVL----FGSVGSRAPRVPNPP 163
Query: 593 PKP----SDGAPVSNSLNE----IPYAP------------SVPSPPPPPGSLPRGAVGDD 632
PKP S +P S S E IP P + P PPPPP V
Sbjct: 164 PKPTSSSSPSSPSSVSSGETERRIPPPPPPPPRKPASNAAAAPPPPPPPPPAKAVRVAPA 343
Query: 633 KVKRAPELVEFYQSLMKREA 652
KV+R PE+ EFY SLM+R++
Sbjct: 344 KVRRVPEVEEFYHSLMRRDS 403
>TC208480
Length = 1576
Score = 53.1 bits (126), Expect = 5e-07
Identities = 73/285 (25%), Positives = 131/285 (45%), Gaps = 38/285 (13%)
Frame = +3
Query: 69 EEREMKLQSELLEYYSL-----KEQVPVIEEFQRQLRIKSV-EIDMLHM--------TIK 114
E+ +M +Q E + ++ K V V +E Q IK + E + +H+ TIK
Sbjct: 54 EKHDMDVQKEAISEDTIRNLKEKNDVHVQKETLSQETIKKLKEENEVHIQEDFLSKETIK 233
Query: 115 SLQEENNKL------QEELIHEASAKRELEVARN-----KIKELQRQIKIIANQTKGQL- 162
+L+EEN+KL EE+I+ EL+ ++ +I +LQ + + + G +
Sbjct: 234 NLKEENDKLLQKVVSLEEVINNLQTDNELQTQKHTSLEMRIAQLQSENNSLLQKEAGLVE 413
Query: 163 ---LLLKQKVSGLQAKEEEVVKKDAEIENNLKTVNDFEIEKKLKTV------NDLEIEAV 213
LL +KV L K E + +K +EN L + ++ E+ + + N L +
Sbjct: 414 KTNQLLNEKVV-LSLKAESLERKINLLENELSSFSEKEVGLETRIAQLQSENNSLLQKEA 590
Query: 214 GLKRRNKELQHEKRELTVKLNAAESRITELSSVTENEMIADAKSETGRLRHANED---LQ 270
L R +L +EK L++K + E +I L S N ++ S + + N + LQ
Sbjct: 591 TLVERTNQLLNEKAVLSLKGESLEQKIYLLES-DLNSLVKKENSTKETISNLNGNIAVLQ 767
Query: 271 KQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYKAPSGKSLARD 315
QVE L+ +R + E LR + L+ ++N++ + S + D
Sbjct: 768 AQVEELEESRNNLFLENQQLREKVSSLQSTVQNHENSNTSSCSWD 902
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,370,816
Number of Sequences: 63676
Number of extensions: 447222
Number of successful extensions: 9626
Number of sequences better than 10.0: 513
Number of HSP's better than 10.0 without gapping: 4910
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7583
length of query: 915
length of database: 12,639,632
effective HSP length: 106
effective length of query: 809
effective length of database: 5,889,976
effective search space: 4764990584
effective search space used: 4764990584
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146570.7


