

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
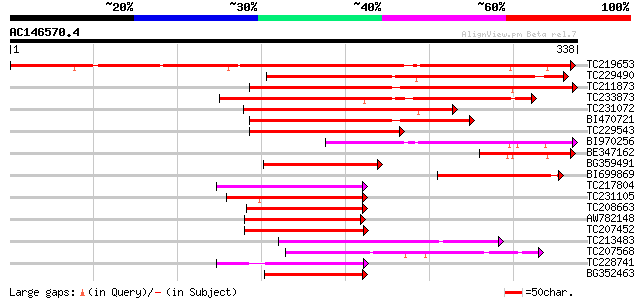
(338 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K1... 437 e-123
TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partia... 174 5e-44
TC211873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein famil... 140 1e-33
TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein famil... 137 9e-33
TC231072 weakly similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622... 124 8e-29
BI470721 109 2e-24
TC229543 similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), part... 103 1e-22
BI970256 85 4e-17
BE347162 83 2e-16
BG359491 77 1e-14
BI699869 66 3e-11
TC217804 63 2e-10
TC231105 62 3e-10
TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacti... 62 4e-10
AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thali... 59 4e-09
TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%) 58 5e-09
TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, parti... 57 1e-08
TC207568 similar to UP|Q9LSL1 (Q9LSL1) Arabidopsis thaliana geno... 57 2e-08
TC228741 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacti... 56 3e-08
BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thalian... 56 3e-08
>TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K13N2_1
{Arabidopsis thaliana;} , partial (47%)
Length = 1346
Score = 437 bits (1124), Expect = e-123
Identities = 251/357 (70%), Positives = 277/357 (77%), Gaps = 20/357 (5%)
Frame = +2
Query: 1 MIQEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNS---DHHIFGGGRSMFPTAGGDHN 57
MIQEDQGQCSSQ INN+ QEQ LLQQQM +QQ ++ IFGGG +M+P GG+ +
Sbjct: 47 MIQEDQGQCSSQAINNYHQAYQEQLLLQQQMQQQQQQQQQNNDIFGGGLNMYP--GGEVS 220
Query: 58 QVSPILQ-QTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLFNRRVPSLQFAYDHHAGS 116
Q+ Q WSM HHHHH HD F++P Q SPYASLFNRR PSLQFAYDH + S
Sbjct: 221 QIMHHHHHQPWSMTMPHHHHHHHQVHDLFLVPPQ-TSPYASLFNRRGPSLQFAYDHGSSS 397
Query: 117 EHLRIISDTLQHG---SGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERI 173
+HLRIIS++ GS PFG Q E+ KM+AQEIMEAKALAASKSHSEAERRRRERI
Sbjct: 398 DHLRIISESFVGPVVQPGSAPFG-LQTELAKMTAQEIMEAKALAASKSHSEAERRRRERI 574
Query: 174 NNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELTVDAAAD 233
NNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTE DELTV AD
Sbjct: 575 NNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTEADELTVVDEAD 754
Query: 234 DEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFITG 293
++ GN +IKASLCC+DRSDL PELIKTLKALRLRTLKA+ITTLGGRVKNVLFITG
Sbjct: 755 ED-----GNS-VIKASLCCEDRSDLFPELIKTLKALRLRTLKAEITTLGGRVKNVLFITG 916
Query: 294 EEDD-----------HEYCISSIQEALKAVMEKSVGD--ESASGSVKRQRTNIISIS 337
EE D + CISSIQEALKAVMEKSVGD ESAS ++KRQRTNIIS+S
Sbjct: 917 EETDSSSSEDHSQQQQQCCISSIQEALKAVMEKSVGDHHESASANIKRQRTNIISMS 1087
>TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partial (56%)
Length = 1006
Score = 174 bits (441), Expect = 5e-44
Identities = 93/182 (51%), Positives = 129/182 (70%), Gaps = 2/182 (1%)
Frame = +1
Query: 154 KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
+ALAA K+H EAE+RRRERIN+HL LR+LLP +KTDKASLLA+V+Q VKELK+QTS I
Sbjct: 322 RALAAMKNHKEAEKRRRERINSHLDHLRTLLPCNSKTDKASLLAKVVQRVKELKQQTSEI 501
Query: 214 AETSPVPTECDELTVDAAADDEDYGSNG--NKFIIKASLCCDDRSDLLPELIKTLKALRL 271
E VP+E DE+TV + DY S G + I KASLCC+DRSDL+P+LI+ L +L L
Sbjct: 502 TELETVPSETDEITV-LSTTGGDYASGGGDGRLIFKASLCCEDRSDLIPDLIEILNSLHL 678
Query: 272 RTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASGSVKRQRT 331
+TLKA++ TLGGR +NVL + +++ I +Q +L++++ D S+SG ++R
Sbjct: 679 KTLKAEMATLGGRTRNVLVVAADKEHSIESIHFLQNSLRSIL-----DRSSSGDRSKRRR 843
Query: 332 NI 333
+
Sbjct: 844 GL 849
>TC211873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein family-like,
partial (25%)
Length = 1149
Score = 140 bits (352), Expect = 1e-33
Identities = 79/202 (39%), Positives = 124/202 (61%), Gaps = 7/202 (3%)
Frame = +3
Query: 144 KMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHV 203
++ + + +++ A KSHSEAER+RR RIN HL LRS++P K DKASLL EVI+H+
Sbjct: 330 RLEKKGVSTERSIEALKSHSEAERKRRARINAHLDTLRSVIPGAMKMDKASLLGEVIRHL 509
Query: 204 KELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELI 263
KELK+ + E +P + DE++V ++++ G NG + I+ASLCC+ + LL ++
Sbjct: 510 KELKKNAAQACEGLMIPKDNDEISV----EEQEGGLNGFPYSIRASLCCEYKPGLLSDIK 677
Query: 264 KTLKALRLRTLKADITTLGGRVKNVLFITGEEDDH-------EYCISSIQEALKAVMEKS 316
+ L AL L +ADI TL GR+KNV I ++ + ++ S+ +ALK+V+ +
Sbjct: 678 QALDALHLMITRADIATLEGRMKNVFVIISCKEQNFEDAAYRQFLAVSVHQALKSVLNRF 857
Query: 317 VGDESASGSVKRQRTNIISISN 338
E G+ KR+R +I S S+
Sbjct: 858 SVSEDILGTRKRRRISIFSSSS 923
>TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein family-like,
partial (25%)
Length = 817
Score = 137 bits (344), Expect = 9e-33
Identities = 79/206 (38%), Positives = 125/206 (60%), Gaps = 17/206 (8%)
Frame = +3
Query: 126 LQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLP 185
L H +G + V A+ E K+ A KSH EAERRRR+RIN+HL+ LR+LLP
Sbjct: 39 LDHDAGHAQSSLIKKAVELDGARSKTERKSTEACKSHREAERRRRQRINSHLSTLRTLLP 218
Query: 186 STTKTDKASLLAEVIQHVKELKRQT-----------------SLIAETSPVPTECDELTV 228
+ K+DKASLL EV++HVK L++Q S+ +E P P ECDE+TV
Sbjct: 219 NAAKSDKASLLGEVVEHVKRLRKQADDVTCGDSYSSRSGEPGSVRSEAWPFPGECDEVTV 398
Query: 229 DAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNV 288
+ D ED G +KA++CC DR+ L ++ + ++++R + ++A++ T+GGR K+V
Sbjct: 399 -SYCDGED----GEPKRVKATVCCGDRTGLNRDVSQAIRSVRAKAVRAEMMTVGGRTKSV 563
Query: 289 LFITGEEDDHEYCISSIQEALKAVME 314
+ + E+++ E + +++ ALKAV+E
Sbjct: 564 VVVEWEKEEEE--VGALERALKAVVE 635
>TC231072 weakly similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), partial
(21%)
Length = 590
Score = 124 bits (310), Expect = 8e-29
Identities = 68/133 (51%), Positives = 87/133 (65%), Gaps = 5/133 (3%)
Frame = +1
Query: 140 GEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEV 199
GE A E +A +ASKSHS+AE+RRR+RIN LA LR L+P + K DKA+LL V
Sbjct: 187 GEFPSWPAPIAAEDRAASASKSHSQAEKRRRDRINAQLATLRKLIPMSDKMDKATLLGSV 366
Query: 200 IQHVKELKRQTSLIAETSPVPTECDELTVD-AAADDEDYGSNGN----KFIIKASLCCDD 254
+ HVK+LKR+ +++ VPTE DE+T+D A DE Y N IIKAS+CCDD
Sbjct: 367 VDHVKDLKRKAMDVSKAITVPTETDEVTIDYHQAQDESYTKKVNILKENIIIKASVCCDD 546
Query: 255 RSDLLPELIKTLK 267
R +L PELI+ LK
Sbjct: 547 RPELFPELIQVLK 585
>BI470721
Length = 421
Score = 109 bits (272), Expect = 2e-24
Identities = 57/134 (42%), Positives = 87/134 (64%)
Frame = +1
Query: 144 KMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHV 203
K+ + + +++ A K+HSEAERRRR RIN HL LRS++P K DKA+LL EVI+H+
Sbjct: 31 KLERKGVSPERSIEALKNHSEAERRRRARINAHLDTLRSVIPGAKKLDKATLLGEVIRHL 210
Query: 204 KELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELI 263
KELK + +E +P + DE+ V ++++ G NG + IKASLCC+ + LL ++
Sbjct: 211 KELKTNATQASEGLMIPKDSDEIRV----EEQEGGLNGFPYSIKASLCCEYKPGLLTDIR 378
Query: 264 KTLKALRLRTLKAD 277
+ L AL L ++A+
Sbjct: 379 QALDALHLMIIRAE 420
>TC229543 similar to UP|Q9LGL7 (Q9LGL7) ESTs C26093(C11622), partial (24%)
Length = 1149
Score = 103 bits (256), Expect = 1e-22
Identities = 54/92 (58%), Positives = 67/92 (72%)
Frame = +2
Query: 144 KMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHV 203
K+ EI EAKALAA K+HSEAERRRRERIN HLA LR L+PST K DKA+LLAEVI V
Sbjct: 836 KVGKSEICEAKALAALKNHSEAERRRRERINGHLATLRGLVPSTEKMDKATLLAEVISQV 1015
Query: 204 KELKRQTSLIAETSPVPTECDELTVDAAADDE 235
KELK+ + +++ +P + DE+ V+ D E
Sbjct: 1016KELKKNAAEVSKGFLIPKDADEVKVEPYNDHE 1111
>BI970256
Length = 703
Score = 85.1 bits (209), Expect = 4e-17
Identities = 62/162 (38%), Positives = 91/162 (55%), Gaps = 12/162 (7%)
Frame = -3
Query: 189 KTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKA 248
K D A LAEVI VKELK+ +++ +P + DE+ V+ D E G G+ A
Sbjct: 695 KMDXAXXLAEVISQVKELKKNXXXVSKGFLIPKDADEVKVEPYNDHE--GGEGS-MSYSA 525
Query: 249 SLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNV-LFITGEED----DHEYC-- 301
++CCD R ++L +L +TL +L L +KA+I+TL GR+KNV +F +E+ D E C
Sbjct: 524 TICCDFRPEILXDLRQTLDSLPLHLVKAEISTLAGRMKNVFVFPCCKENINNIDFEKCQA 345
Query: 302 -ISSIQEALKAVMEKSVG----DESASGSVKRQRTNIISISN 338
SS+ +AL +VMEK+ S + KR+R I SN
Sbjct: 344 LASSVPQALCSVMEKASASLDFSPRTSHASKRRRLCFIETSN 219
>BE347162
Length = 424
Score = 83.2 bits (204), Expect = 2e-16
Identities = 50/77 (64%), Positives = 56/77 (71%), Gaps = 20/77 (25%)
Frame = +2
Query: 281 LGGRVKNVLFITGEE--------DDH----------EYCISSIQEALKAVMEKSVGD--E 320
LGGRVKNVLFITGEE +DH +YCI+SIQEALKAVMEKSVGD E
Sbjct: 2 LGGRVKNVLFITGEEADSSSGSTEDHSHHHHQQQQQQYCINSIQEALKAVMEKSVGDHHE 181
Query: 321 SASGSVKRQRTNIISIS 337
SAS ++KRQRTNIIS+S
Sbjct: 182 SASANIKRQRTNIISMS 232
>BG359491
Length = 361
Score = 77.0 bits (188), Expect = 1e-14
Identities = 38/71 (53%), Positives = 51/71 (71%)
Frame = +2
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTS 211
E +A++ASKSH +AE+RRR+RIN LA LR L+P + K DKA+LL VI VK+LKR+
Sbjct: 146 EERAISASKSHRQAEKRRRDRINAQLATLRKLIPKSDKMDKAALLGSVIDQVKDLKRKAM 325
Query: 212 LIAETSPVPTE 222
++ VPTE
Sbjct: 326 DVSRAFTVPTE 358
>BI699869
Length = 421
Score = 65.9 bits (159), Expect = 3e-11
Identities = 34/75 (45%), Positives = 52/75 (69%)
Frame = +1
Query: 256 SDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEK 315
SDL+PELI+ L++LRL+TLKA++ TLGGR +NVL + ++D I +Q +LK+++E+
Sbjct: 1 SDLIPELIEILRSLRLKTLKAEMATLGGRTRNVLVVATDKDHSGESIQFLQNSLKSLVER 180
Query: 316 SVGDESASGSVKRQR 330
S S KR+R
Sbjct: 181 SSNSNDRS---KRRR 216
>TC217804
Length = 1578
Score = 63.2 bits (152), Expect = 2e-10
Identities = 31/90 (34%), Positives = 54/90 (59%)
Frame = +1
Query: 124 DTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSL 183
D++ H G + K+ + A S H ++ERRRR++IN + +L+ L
Sbjct: 709 DSISHRISQGEVPDEDYKATKVDRSSGSNKRIKANSVVHKQSERRRRDKINQRMKELQKL 888
Query: 184 LPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
+P+++KTDKAS+L EVIQ++K+L+ Q ++
Sbjct: 889 VPNSSKTDKASMLDEVIQYMKQLQAQVQMM 978
>TC231105
Length = 449
Score = 62.4 bits (150), Expect = 3e-10
Identities = 33/86 (38%), Positives = 56/86 (64%), Gaps = 2/86 (2%)
Frame = +2
Query: 130 SGSGPFGGFQGEVGKMSA--QEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPST 187
S S P G Q E K A + + K A+ H+++ER+RR++IN + L+ L+P++
Sbjct: 83 SHSKPMGEDQDEEKKKRANGKSSVSTKRSRAAAIHNQSERKRRDKINQRMKTLQKLVPNS 262
Query: 188 TKTDKASLLAEVIQHVKELKRQTSLI 213
+K+DKAS+L EVI+++K+L+ Q +I
Sbjct: 263 SKSDKASMLDEVIEYLKQLQAQLQMI 340
>TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4
(Basic helix-loop-helix protein 9) (bHLH9) (AtbHLH009)
(Short under red-light 2), partial (19%)
Length = 1102
Score = 62.0 bits (149), Expect = 4e-10
Identities = 30/72 (41%), Positives = 49/72 (67%)
Frame = +1
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G S+Q A+ A++ H+ +ERRRR+RIN + L+ L+P ++KTDKAS+L E I+
Sbjct: 214 LGNKSSQRTGSARRNRAAEVHNLSERRRRDRINEKMKALQQLIPHSSKTDKASMLEEAIE 393
Query: 202 HVKELKRQTSLI 213
++K L+ Q L+
Sbjct: 394 YLKSLQLQLQLM 429
>AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thaliana},
partial (17%)
Length = 381
Score = 58.5 bits (140), Expect = 4e-09
Identities = 28/72 (38%), Positives = 48/72 (65%)
Frame = +1
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
EV +A +K A++ H+ +E+RRR RIN + L++L+P++ KTDKAS+L E I
Sbjct: 166 EVPTKAASSRSSSKRSRAAEVHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAI 345
Query: 201 QHVKELKRQTSL 212
+++K+L+ Q +
Sbjct: 346 EYLKQLQLQVQM 381
>TC207452 similar to UP|Q9FUA4 (Q9FUA4) SPATULA, partial (31%)
Length = 1207
Score = 58.2 bits (139), Expect = 5e-09
Identities = 28/74 (37%), Positives = 50/74 (66%)
Frame = +2
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
EV +A +K A++ H+ +E+RRR RIN + L++L+P++ KTDKAS+L E I
Sbjct: 101 EVPTKAASARSSSKRSRAAEVHN*SEKRRRGRINEKMKALQNLIPNSNKTDKASMLDEAI 280
Query: 201 QHVKELKRQTSLIA 214
+++K+L+ Q +++
Sbjct: 281 EYLKQLQLQVQMLS 322
>TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, partial (17%)
Length = 914
Score = 57.0 bits (136), Expect = 1e-08
Identities = 33/134 (24%), Positives = 68/134 (50%)
Frame = +2
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP 220
+H EAER+RRE++N LRS++P+ +K DKASLL + I ++ EL+ + ++
Sbjct: 155 NHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDTIAYINELQAKVKIMEAERERF 334
Query: 221 TECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITT 280
+A AD + ++ I++ S D+ L ++I+T ++ +++ + +
Sbjct: 335 ESISNQEKEAPADVDIQAVQDDEVIVRVSCPLDNHP--LSKVIQTFYQTQISVVESKLAS 508
Query: 281 LGGRVKNVLFITGE 294
+ + I +
Sbjct: 509 ANDAIFHTFVIKSQ 550
>TC207568 similar to UP|Q9LSL1 (Q9LSL1) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K21L13, partial (50%)
Length = 914
Score = 56.6 bits (135), Expect = 2e-08
Identities = 44/165 (26%), Positives = 82/165 (49%), Gaps = 11/165 (6%)
Frame = +1
Query: 165 AERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECD 224
AERRRR+R+N+ L+ LRS++P +K D+ S+L + I ++KEL + + E + +
Sbjct: 142 AERRRRKRLNDRLSMLRSIVPKISKMDRTSILGDTIDYMKELLERIGKLQE-EEIEEGTN 318
Query: 225 ELTVDAAADD----EDYGSNGNKFII-------KASLCCDDRSDLLPELIKTLKALRLRT 273
++ + + + E N KF + + S+CC + LL + TL+AL L
Sbjct: 319 QINLLGISKELKPNEVMVRNSPKFDVERRDQDTRISICCATKPGLLLSTVNTLEALGLEI 498
Query: 274 LKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVG 318
+ I++ ++ E + C+S QE +K + ++ G
Sbjct: 499 HQCVISSFND--FSMQASCTEVAEQRNCMS--QEEIKQALFRNAG 621
>TC228741 similar to UP|PIF3_ARATH (O80536) Phytochrome-interacting factor 3
(Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008), partial
(25%)
Length = 763
Score = 55.8 bits (133), Expect = 3e-08
Identities = 30/91 (32%), Positives = 52/91 (56%)
Frame = +3
Query: 124 DTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSL 183
D + +G+ G QG G +K A++ H+ +ER+RR+RIN + L+ L
Sbjct: 222 DVEEESAGAKKTAGGQGGAG---------SKRSRAAEVHNLSERKRRDRINEKMRALQEL 374
Query: 184 LPSTTKTDKASLLAEVIQHVKELKRQTSLIA 214
+P+ K DKAS+L E I+++K L+ Q +++
Sbjct: 375 IPNCNKVDKASMLDEAIEYLKTLQLQVQIMS 467
>BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thaliana}, partial
(19%)
Length = 471
Score = 55.8 bits (133), Expect = 3e-08
Identities = 25/61 (40%), Positives = 44/61 (71%)
Frame = +3
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSL 212
+K A++ H+ +E+RRR RIN + L++L+P++ KTDKAS+L E I+++K+L+ Q
Sbjct: 36 SKRSRAAEFHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQY 215
Query: 213 I 213
+
Sbjct: 216 L 218
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,730,354
Number of Sequences: 63676
Number of extensions: 227210
Number of successful extensions: 3297
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 2735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3098
length of query: 338
length of database: 12,639,632
effective HSP length: 98
effective length of query: 240
effective length of database: 6,399,384
effective search space: 1535852160
effective search space used: 1535852160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146570.4


