

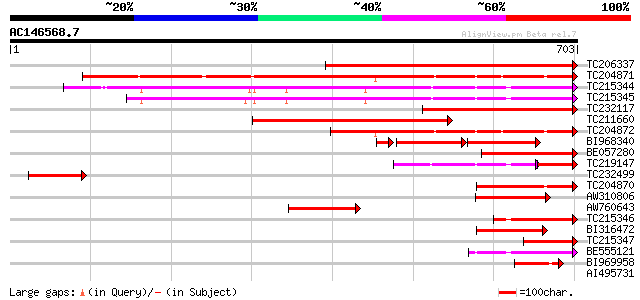
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.7 - phase: 0
(703 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), par... 522 e-148
TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%) 474 e-134
TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 403 e-112
TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 363 e-100
TC232117 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), par... 350 1e-96
TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), par... 336 2e-92
TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%) 267 1e-71
BI968340 134 2e-65
BE057280 similar to GP|9295728|gb| T24P13.25 {Arabidopsis thalia... 161 1e-39
TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11... 110 1e-31
TC232499 similar to UP|Q9ZVH4 (Q9ZVH4) T2P11.2 protein (T24P13.2... 124 1e-28
TC204870 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (17%) 114 2e-25
AW310806 95 1e-19
AW760643 86 6e-17
TC215346 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 79 7e-15
BI316472 75 1e-13
TC215347 homologue to UP|Q711G6 (Q711G6) Cullin 1C (Fragment), p... 68 2e-11
BE555121 similar to GP|22335691|db cullin-like protein1 {Pisum s... 67 2e-11
BI969958 45 1e-04
AI495731 weakly similar to GP|23429518|gb APC2 {Arabidopsis thal... 40 0.004
>TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), partial (91%)
Length = 1135
Score = 522 bits (1344), Expect = e-148
Identities = 261/313 (83%), Positives = 282/313 (89%), Gaps = 1/313 (0%)
Frame = +3
Query: 392 KYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYA 451
KYYKQHLAKRLLSGKT+SDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTS DTMQGFYA
Sbjct: 3 KYYKQHLAKRLLSGKTISDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSHDTMQGFYA 182
Query: 452 S-HPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQT 510
+LGDGP L+VQVLTTGSWPTQ S CNLPVEI +C+KFR+YYLGTH GRRLSWQT
Sbjct: 183 ILGTELGDGPLLSVQVLTTGSWPTQPSPPCNLPVEILGVCDKFRTYYLGTHNGRRLSWQT 362
Query: 511 NMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQS 570
NMG ADLKATFGKGQKHELNVSTYQMCVLMLFN+A++L+ KEIEQAT IP DL+RCLQS
Sbjct: 363 NMGTADLKATFGKGQKHELNVSTYQMCVLMLFNSAERLTCKEIEQATAIPMSDLRRCLQS 542
Query: 571 LALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRV 630
LA VKG+NVLRKEPMSKD+ EDDAF NDKF+SK +KVKIGTVVAQ+ESEPE ETRQRV
Sbjct: 543 LACVKGKNVLRKEPMSKDIAEDDAFFFNDKFTSKFFKVKIGTVVAQRESEPENLETRQRV 722
Query: 631 EEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLE 690
EEDRKPQIEAAIVRIMKSRR LDHNN++AEVTKQLQ RFL NP +KKRIESLIER+FLE
Sbjct: 723 EEDRKPQIEAAIVRIMKSRRTLDHNNIVAEVTKQLQSRFLPNPVVIKKRIESLIEREFLE 902
Query: 691 RDDNDRKMYRYLA 703
RD DRK+YRYLA
Sbjct: 903 RDKVDRKLYRYLA 941
>TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%)
Length = 2087
Score = 474 bits (1221), Expect = e-134
Identities = 265/618 (42%), Positives = 405/618 (64%), Gaps = 5/618 (0%)
Frame = +1
Query: 91 MDRTFIP-SAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMR 149
+DRT++ +A + ++GL L+R+ + S ++ + + LL +++SER GE +DR ++
Sbjct: 1 LDRTYVKQTANVRSLWDMGLQLFRKHLSLSPEVEHKTVTGLLRMIESERKGEAVDRTLLN 180
Query: 150 NITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDR 209
++ KM LG +Y + FE FL+ ++EFY E ++++ D DYLK E RL EE +R
Sbjct: 181 HLLKMFTALG--IYAESFEKPFLECTSEFYAAEGVKYMQQSDVPDYLKHVEIRLQEEHER 354
Query: 210 VGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVA 269
Y+D T K + E Q++E H+ ++ + G ++ ++ EDL RMY+LF RV
Sbjct: 355 CLIYLDASTRKPLIATAEKQLLERHIPAIL---DKGFAMLMDGNRIEDLQRMYSLFSRV- 522
Query: 270 DGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQ 329
+ L +R+ ++ +IR +G+ +V D E+ KD V LL+ K D ++F+ +++F
Sbjct: 523 NALESLRQAISSYIRRTGQGIVLDEEKDKD---MVSSLLEFKASLDTTWEESFSKNEAFC 693
Query: 330 NALNSSFEYFINLNPRSP-EFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKD 388
N + SFE+ INL P E I+ F+D+KLR G KG +E+++E TLDKV++LFR++Q KD
Sbjct: 694 NTIKDSFEHLINLRQNRPAELIAKFLDEKLRAGNKGTSEEELEGTLDKVLVLFRFIQGKD 873
Query: 389 VFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQG 448
VFE +YK+ LAKRLL GK+ S DAE+S+I KLKTECG QFT+KLEGMF D++ S++ +
Sbjct: 874 VFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKDIELSKEINES 1053
Query: 449 FYAS---HPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRR 505
F S L G ++V VLTTG WPT + LP E++ + F+ +YL ++GRR
Sbjct: 1054FKQSSQARTKLPSGIEMSVHVLTTGYWPTYPPMDVRLPHELNVYQDIFKEFYLSKYSGRR 1233
Query: 506 LSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLK 565
L WQ ++G LKA F KG+K EL VS +Q VLMLFN+A+KLS+++I+ +T I +L+
Sbjct: 1234LMWQNSLGHCVLKAEFPKGKK-ELAVSLFQTVVLMLFNDAEKLSFQDIKDSTGIEDKELR 1410
Query: 566 RCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQE 625
R LQSLA K R VL+K P +DV +DD+F N+ F++ LY++K+ + KE+ E
Sbjct: 1411RTLQSLACGKVR-VLQKLPKGRDVEDDDSFVFNEGFTAPLYRIKV-NAIQLKETVEENTS 1584
Query: 626 TRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIE 685
T +RV +DR+ Q++AAIVRIMK+R++L H LI E+ + QL+F P ++KKRIESLI+
Sbjct: 1585TTERVFQDRQYQVDAAIVRIMKTRKVLSHTLLITELFQ--QLKFPIKPADLKKRIESLID 1758
Query: 686 RDFLERDDNDRKMYRYLA 703
R++LERD N+ ++Y YLA
Sbjct: 1759REYLERDKNNPQIYNYLA 1812
>TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (92%)
Length = 2465
Score = 403 bits (1035), Expect = e-112
Identities = 235/660 (35%), Positives = 380/660 (56%), Gaps = 23/660 (3%)
Frame = +1
Query: 67 LEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRL 126
L EL ++W +H ++ + Y+DR FI P++E+GL +R+ +IY ++ ++
Sbjct: 100 LRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLTCFRD-LIYK-ELNGKV 273
Query: 127 LNTLLELVQSERTGEVIDRGIMRNITKMLMDLGPAV---YGQDFEAHFLQVSAEFYQVES 183
+ ++ L+ ER GE IDR +++N+ + +++G Y DFEA L+ ++ +Y ++
Sbjct: 274 RDAVISLIDQEREGEQIDRALLKNVLDIFVEIGMGQMDHYENDFEAAMLKDTSAYYSRKA 453
Query: 184 QRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMEN 243
+I C DY+ KAE L E DRV HY+ +E K+ + V+ +++ + +L+ E+
Sbjct: 454 SNWILEDSCPDYMLKAEECL*REKDRVAHYLHSSSEPKLLEKVQHELLSVYANQLLEKEH 633
Query: 244 SGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPE-------- 295
SG +L DDK EDL RM+ LF ++ GL + + H+ G LV E
Sbjct: 634 SGCHALLRDDKVEDLSRMFRLFSKIPRGLDPVSNIFKQHVTTEGMALVKQAEDAASNKKA 813
Query: 296 RLKDPVE-----FVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSP 347
KD V FV+++++ DKY +N F N F AL +FE F N S
Sbjct: 814 EKKDIVGLQEQVFVRKVIELHDKYLAYVNDCFQNHTLFHKALKEAFEVFCNKGVAGSSSA 993
Query: 348 EFISLFVDDKLRKG-LKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGK 406
E ++ F D+ L+KG + ++++ +E TL+KV+ L Y+ +KD+F ++Y++ LA+RLL K
Sbjct: 994 ELLASFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDK 1173
Query: 407 TVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM---KTSQDTMQGFYASHPDLGDGPTLT 463
+ +DD ERS++ KLK +CG QFTSK+EGM TD+ K +Q + + + +++P+ G LT
Sbjct: 1174SANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNNPNADPGIDLT 1353
Query: 464 VQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGK 523
V VLTTG WP+ S NLP E+ E F+ +Y R+L+W ++G ++ F
Sbjct: 1354VTVLTTGFWPSYKSFDLNLPAEMIRCVEVFKEFYQTKTKHRKLTWIYSLGTCNISGKFDP 1533
Query: 524 GQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKE 583
+ EL V+TYQ L+LFN++D+LSY EI + D+ R L SL+ K + +L KE
Sbjct: 1534-KTVELIVTTYQASALLLFNSSDRLSYSEIMTQLNLSDDDVIRLLHSLSCAKYK-ILNKE 1707
Query: 584 PMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIV 643
P +K + D F N KF+ K+ ++KI EK++ + V++DR+ I+A+IV
Sbjct: 1708PNTKTISSTDYFEFNYKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIV 1872
Query: 644 RIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
RIMKSR++L + L+ E +QL F + +KKRIE LI RD+LERD ++ M++YLA
Sbjct: 1873RIMKSRKVLGYQQLVVECVEQLGRMFKPDVKAIKKRIEDLISRDYLERDKDNANMFKYLA 2052
>TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (77%)
Length = 2106
Score = 363 bits (932), Expect = e-100
Identities = 211/582 (36%), Positives = 335/582 (57%), Gaps = 23/582 (3%)
Frame = +1
Query: 145 RGIMRNITKMLMDLGPAV---YGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAER 201
R +++N+ + +++G Y DFEA L+ ++ +Y ++ +I C DY+ KAE
Sbjct: 1 RALLKNVLDIFVEIGMGQMDHYENDFEAAMLKDTSAYYSRKASNWILEDSCPDYMLKAEE 180
Query: 202 RLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRM 261
L E DRV HY+ +E K+ + V+ +++ + +L+ E+SG +L DDK EDL RM
Sbjct: 181 CLKREKDRVAHYLHSSSEPKLLEKVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLSRM 360
Query: 262 YNLFRRVADGLLKIREVMTLHIRESGKQLV--------TDPERLKDPVE-----FVQRLL 308
+ LF ++ GL + + H+ G LV T KD V FV++++
Sbjct: 361 FRLFSKIPRGLDPVSSIFKQHVTAEGMALVKLAEDAVSTKKAEKKDIVGLQEQVFVRKVI 540
Query: 309 DEKDKYDKIINQAFNNDKSFQNALNSSFEYFIN---LNPRSPEFISLFVDDKLRKG-LKG 364
+ DKY +N F N F AL +FE F N S E ++ F D+ L+KG +
Sbjct: 541 ELHDKYLAYVNDCFQNHTLFHKALKEAFEIFCNKGVAGSSSAELLATFCDNILKKGGSEK 720
Query: 365 VNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTEC 424
++++ +E TL+KV+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ERS++ KLK +C
Sbjct: 721 LSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQC 900
Query: 425 GYQFTSKLEGMFTDM---KTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCN 481
G QFTSK+EGM TD+ K +Q + + + ++P+ G LTV VLTTG WP+ S N
Sbjct: 901 GGQFTSKMEGMVTDLTLAKENQTSFEEYLTNNPNADPGIDLTVTVLTTGFWPSYKSFDLN 1080
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLML 541
LP E+ E F+ +Y R+L+W ++G ++ F + EL V+TYQ L+L
Sbjct: 1081LPAEMVRCVEVFKEFYQTKTKHRKLTWIYSLGTCNISGKFDP-KTVELIVTTYQASALLL 1257
Query: 542 FNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKF 601
FN++D+LSY EI + D+ R L SL+ K + +L KEP +K + D F N KF
Sbjct: 1258FNSSDRLSYSEIMTQLNLSDDDVIRLLHSLSCAKYK-ILNKEPNTKTISSTDYFEFNSKF 1434
Query: 602 SSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEV 661
+ K+ ++KI EK++ + V++DR+ I+A+IVRIMKSR++L++ L+ E
Sbjct: 1435TDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLNYQQLVMEC 1599
Query: 662 TKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+QL F + +KKRIE LI RD+LERD ++ ++RYLA
Sbjct: 1600VEQLGRMFKPDVKAIKKRIEDLISRDYLERDKDNANLFRYLA 1725
>TC232117 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), partial (57%)
Length = 832
Score = 350 bits (898), Expect = 1e-96
Identities = 177/192 (92%), Positives = 184/192 (95%)
Frame = +1
Query: 512 MGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSL 571
MG ADLKATFGKGQKHELNVSTYQMCVLMLFNNAD+ YKEIEQATEIPA DLKRCLQSL
Sbjct: 1 MGTADLKATFGKGQKHELNVSTYQMCVLMLFNNADRXXYKEIEQATEIPASDLKRCLQSL 180
Query: 572 ALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVE 631
ALVKGRNVLRKEPM KD+G+DDAF VNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVE
Sbjct: 181 ALVKGRNVLRKEPMGKDIGDDDAFYVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVE 360
Query: 632 EDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLER 691
EDRKPQIEAAIVRIMKSR+ LDHNNLIAEVTKQLQ RFLANPTEVKKRIESLIERDFLER
Sbjct: 361 EDRKPQIEAAIVRIMKSRKQLDHNNLIAEVTKQLQSRFLANPTEVKKRIESLIERDFLER 540
Query: 692 DDNDRKMYRYLA 703
DD+DR++YRYLA
Sbjct: 541 DDSDRRLYRYLA 576
>TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), partial (29%)
Length = 747
Score = 336 bits (862), Expect = 2e-92
Identities = 171/249 (68%), Positives = 203/249 (80%), Gaps = 1/249 (0%)
Frame = +1
Query: 302 EFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKG 361
+FVQRLLD KDKYD++I +FNNDK+FQNALNSSFEYFINLN RSPEFISLFVDDKLR+G
Sbjct: 1 DFVQRLLDLKDKYDRVITMSFNNDKTFQNALNSSFEYFINLNARSPEFISLFVDDKLRRG 180
Query: 362 LKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLK 421
LKGV E+DVE+ L+KVMMLFRYLQEKDVFEKYYKQHLAKRLLS KT+S DAERSLIVKL+
Sbjct: 181 LKGVGEEDVEIVLNKVMMLFRYLQEKDVFEKYYKQHLAKRLLSRKTISYDAERSLIVKLE 360
Query: 422 TECGYQFTSKLEGMFTDMKTSQDTMQGFYAS-HPDLGDGPTLTVQVLTTGSWPTQSSITC 480
T+CGYQFTSKL+GMFTDM TS DTMQG YA+ +LGDGP L+VQVLTTGSWPTQ + C
Sbjct: 361 TQCGYQFTSKLQGMFTDMTTSHDTMQGLYANLGSELGDGPMLSVQVLTTGSWPTQPTPPC 540
Query: 481 NLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLM 540
NL V I + +K R+YYL +H GRRL TN+ A +K T Q +E + Y M VL+
Sbjct: 541 NLTV*ILRVSDKSRTYYLDSHNGRRLYSLTNIETA*VKTTLCNSQAYEFH*FVYGMSVLV 720
Query: 541 LFNNADKLS 549
L+ + ++++
Sbjct: 721 LYCSIERVT 747
>TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%)
Length = 1224
Score = 267 bits (682), Expect = 1e-71
Identities = 151/309 (48%), Positives = 209/309 (66%), Gaps = 3/309 (0%)
Frame = +2
Query: 398 LAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYAS---HP 454
L +RLL GK+ S DAE+S+I KLKTECG QFT+KLEGMF D++ S++ F S
Sbjct: 26 LLRRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKDIELSKEINDSFKQSSQARS 205
Query: 455 DLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGF 514
L G ++V VLTTG WPT + LP E++ + F+ +YL ++GRRL WQ ++G
Sbjct: 206 KLASGIEMSVHVLTTGHWPTYPPMDVRLPHELNVYQDIFKEFYLSKYSGRRLMWQNSLGH 385
Query: 515 ADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALV 574
LKA F KG+K EL VS +Q VLMLFN+A+KLS ++I+ AT I +L+R LQSLA
Sbjct: 386 CVLKAEFPKGRK-ELAVSLFQTVVLMLFNDAEKLSLQDIKDATGIEDKELRRTLQSLACG 562
Query: 575 KGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDR 634
K R VL+K P +DV +DD F ND F++ LY++K+ + KE+ E T +RV DR
Sbjct: 563 KVR-VLQKMPKGRDVEDDDLFVFNDGFTAPLYRIKV-NAIQLKETVEENTSTTERVFHDR 736
Query: 635 KPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDN 694
+ QI+AAIVRIMK+R++L H LI E+ + QL+F P ++KKRIESLI+R++LERD +
Sbjct: 737 QYQIDAAIVRIMKTRKVLSHTLLITELFQ--QLKFPIKPADLKKRIESLIDREYLERDKS 910
Query: 695 DRKMYRYLA 703
+ ++Y YLA
Sbjct: 911 NPQIYNYLA 937
>BI968340
Length = 621
Score = 134 bits (337), Expect(3) = 2e-65
Identities = 62/87 (71%), Positives = 72/87 (82%)
Frame = -2
Query: 480 CNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVL 539
CNLPVE+ +C+KF +YYLG+H GRRLSWQ NMG DLKA FGKGQKHELNVST QMCVL
Sbjct: 536 CNLPVEMLGVCDKFGTYYLGSHNGRRLSWQGNMGGGDLKAAFGKGQKHELNVSTCQMCVL 357
Query: 540 MLFNNADKLSYKEIEQATEIPAPDLKR 566
MLFN+ ++L+ KEIEQ T IP DL+R
Sbjct: 356 MLFNSXERLTCKEIEQGTAIPMSDLRR 276
Score = 125 bits (313), Expect(3) = 2e-65
Identities = 64/91 (70%), Positives = 74/91 (80%)
Frame = -1
Query: 568 LQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETR 627
LQ +A VKG+NVLRKEPMSKD+ EDDA N KF+SKL+ VKIGTVVAQ+ESEPE ETR
Sbjct: 273 LQCVACVKGKNVLRKEPMSKDIXEDDALFFNGKFASKLFXVKIGTVVAQRESEPENLETR 94
Query: 628 QRVEEDRKPQIEAAIVRIMKSRRLLDHNNLI 658
QRVEEDRKPQI ++IV +KS R L HN L+
Sbjct: 93 QRVEEDRKPQIXSSIVXXIKSXRTLYHNILL 1
Score = 30.0 bits (66), Expect(3) = 2e-65
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = -1
Query: 455 DLGDGPTLTVQVLTTGSWPTQ 475
+LGDG ++VQ LTTGSW Q
Sbjct: 612 ELGDGRVVSVQGLTTGSWGGQ 550
>BE057280 similar to GP|9295728|gb| T24P13.25 {Arabidopsis thaliana}, partial
(16%)
Length = 403
Score = 161 bits (407), Expect = 1e-39
Identities = 83/118 (70%), Positives = 96/118 (81%)
Frame = +3
Query: 586 SKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRI 645
SKD+ EDDAF NDK +SK+ +VKIG+V + ESEPE T Q E+R+PQIEAAIVRI
Sbjct: 15 SKDIAEDDAFFWNDKVTSKVCEVKIGSVGGEXESEPENVGTLQIXGEEREPQIEAAIVRI 194
Query: 646 MKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
MKSRR LDHNN++AEVTKQLQ RFL NP +KKRIESLIER+FLERD DRK+YRYLA
Sbjct: 195 MKSRRTLDHNNIVAEVTKQLQSRFLPNPVVIKKRIESLIEREFLERDKVDRKLYRYLA 368
>TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11.14.1 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(33%)
Length = 1237
Score = 110 bits (276), Expect(2) = 1e-31
Identities = 68/182 (37%), Positives = 103/182 (56%)
Frame = +3
Query: 477 SITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQM 536
S NLP E+ E F+ +Y R+L+W ++G + F + + EL V TY
Sbjct: 9 SFDLNLPSEMIRCLEVFKGFYETRTKHRKLTWIYSLGTCHVTGKF-ETKNIELIVPTYPA 185
Query: 537 CVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFS 596
L+LFNNAD+LSY EI + D+ R L SL+ K + +L KEP +K + + D F
Sbjct: 186 AALLLFNNADRLSYSEIMTQLNLGHEDVARLLHSLSSAKYK-ILIKEPNNKVISQSDIFE 362
Query: 597 VNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNN 656
N KF+ K+ ++KI A E+++ + V++DR+ I+AAIVRIMKSR++L H
Sbjct: 363 FNYKFTDKMRRIKIPLPPAD-----ERKKVIEDVDKDRRYAIDAAIVRIMKSRKILGHQQ 527
Query: 657 LI 658
L+
Sbjct: 528 LV 533
Score = 44.7 bits (104), Expect(2) = 1e-31
Identities = 23/49 (46%), Positives = 30/49 (60%)
Frame = +1
Query: 655 NNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
NN E +QL F + +KKRIE LI RD+LERD ++ +RYLA
Sbjct: 523 NN*FLECVEQLGRMFKPDIKAIKKRIEDLITRDYLERDKDNPNTFRYLA 669
>TC232499 similar to UP|Q9ZVH4 (Q9ZVH4) T2P11.2 protein (T24P13.25) (Cullin
3a), partial (17%)
Length = 432
Score = 124 bits (311), Expect = 1e-28
Identities = 58/72 (80%), Positives = 67/72 (92%)
Frame = +3
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+RNAYNMVL KFG++LY+GLV TMT+HLKEI++SIE+AQG FLEE+NRKW DHNKALQM
Sbjct: 216 YRNAYNMVLQKFGEKLYTGLVTTMTSHLKEISQSIESAQGEIFLEEINRKWVDHNKALQM 395
Query: 84 IRDILMYMDRTF 95
IRDILMYMDRTF
Sbjct: 396 IRDILMYMDRTF 431
>TC204870 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (17%)
Length = 621
Score = 114 bits (284), Expect = 2e-25
Identities = 59/125 (47%), Positives = 89/125 (71%)
Frame = +2
Query: 579 VLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQI 638
VL K P +DV +DD+F N+ F++ LY++K+ + KE+ E T +RV +DR+ Q+
Sbjct: 14 VLPKLPKGRDVEDDDSFVFNEGFTAPLYRIKVNAIQL-KETVEENTSTTERVFQDRQYQV 190
Query: 639 EAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKM 698
+AAIVRIMK+R++L H LI E+ + QL+F P ++KKRIESLI+R++LERD N+ ++
Sbjct: 191 DAAIVRIMKTRKVLSHTLLITELFQ--QLKFPIKPADLKKRIESLIDREYLERDKNNPQI 364
Query: 699 YRYLA 703
Y YLA
Sbjct: 365 YNYLA 379
>AW310806
Length = 301
Score = 94.7 bits (234), Expect = 1e-19
Identities = 51/93 (54%), Positives = 63/93 (66%)
Frame = -1
Query: 578 NVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQ 637
+VLR PMSK + EDD F N +F+ K +KVKIGTVVA ++S+ E R RV+ DR
Sbjct: 301 HVLRPHPMSKPIAEDDVFFFNHQFTXKFFKVKIGTVVA*RQSDLENLALRHRVQPDRHS* 122
Query: 638 IEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFL 670
IE IVRIMKSRR LD+NN++ EV K L FL
Sbjct: 121 IEVVIVRIMKSRRTLDYNNIVXEVIK*L*SWFL 23
>AW760643
Length = 286
Score = 85.9 bits (211), Expect = 6e-17
Identities = 40/91 (43%), Positives = 66/91 (71%), Gaps = 1/91 (1%)
Frame = +1
Query: 346 SPEFISLFVDDKLRKG-LKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLS 404
S E +S F D+ L+KG + ++++ +E TL+KV+ L Y+ +KD+F ++Y+ LA+RLL
Sbjct: 7 SAELLSTFCDNILKKGGSEKLSDEAIEDTLEKVVKLLAYISDKDLFAEFYRNKLARRLLC 186
Query: 405 GKTVSDDAERSLIVKLKTECGYQFTSKLEGM 435
++ +DD E+ ++ KLK +CG QFTSK+EGM
Sbjct: 187 DRSANDDHEKCILTKLKQQCGGQFTSKMEGM 279
>TC215346 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (13%)
Length = 1050
Score = 79.0 bits (193), Expect = 7e-15
Identities = 42/103 (40%), Positives = 66/103 (63%)
Frame = +1
Query: 601 FSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAE 660
F+ K+ ++KI EK++ + V++DR+ I+A+IVRIMKSR++L + L+ E
Sbjct: 1 FTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLSYQQLVME 165
Query: 661 VTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+QL F + +KKRIE LI RD+LERD ++ ++RYLA
Sbjct: 166 CVEQLGRMFKPDVKAIKKRIEDLISRDYLERDKDNANLFRYLA 294
>BI316472
Length = 415
Score = 75.1 bits (183), Expect = 1e-13
Identities = 38/87 (43%), Positives = 59/87 (67%)
Frame = +1
Query: 580 LRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIE 639
+++ P +DV +DD F ND F++ LY++K+ + KE+ E T +RV DR+ QI+
Sbjct: 151 MQQMPKGRDVEDDDLFVFNDGFTAPLYRIKVNAIQL-KETVEENTSTTERVFHDRQYQID 327
Query: 640 AAIVRIMKSRRLLDHNNLIAEVTKQLQ 666
AAIVRIMK+R++L H LI E+ +QL+
Sbjct: 328 AAIVRIMKTRKVLSHTLLITELFQQLK 408
>TC215347 homologue to UP|Q711G6 (Q711G6) Cullin 1C (Fragment), partial (15%)
Length = 668
Score = 67.8 bits (164), Expect = 2e-11
Identities = 33/66 (50%), Positives = 47/66 (71%)
Frame = +1
Query: 638 IEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRK 697
I+A+IVRIMKSR++L + L+ E +QL F + +KKRIE LI RD+LERD ++
Sbjct: 55 IDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAIKKRIEDLISRDYLERDKDNAN 234
Query: 698 MYRYLA 703
M++YLA
Sbjct: 235 MFKYLA 252
>BE555121 similar to GP|22335691|db cullin-like protein1 {Pisum sativum},
partial (17%)
Length = 388
Score = 67.4 bits (163), Expect = 2e-11
Identities = 44/134 (32%), Positives = 71/134 (52%)
Frame = +2
Query: 570 SLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQR 629
SL+ K + +L KEP +K + D N KF+ K+ ++KI EK++ +
Sbjct: 5 SLSCAKNK-ILNKEPNTKTMSYTDYSEYNPKFTDKMRRIKIPFPPVD-----EKKKLIED 166
Query: 630 VEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFL 689
+ R+ ++A IVR++ SR++L H L+ E +QL F + +KK I LI RD L
Sbjct: 167 ADNYRRYAVDATIVRLINSRKVLSHQQLVMECVEQLGHMFKPDVIAIKKLIADLIYRDCL 346
Query: 690 ERDDNDRKMYRYLA 703
D N+ M++ LA
Sbjct: 347 *IDKNNAYMFQDLA 388
>BI969958
Length = 181
Score = 45.1 bits (105), Expect = 1e-04
Identities = 27/61 (44%), Positives = 40/61 (65%)
Frame = -2
Query: 626 TRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIE 685
T +RV DR+ QI+A IV IMK +++ +E+ +QL+L P ++KKRIESLI+
Sbjct: 177 TTERVFHDRQYQIDAGIVXIMKPSKVVSLAVXSSELFQQLKLPI--KPDDLKKRIESLID 4
Query: 686 R 686
R
Sbjct: 3 R 1
>AI495731 weakly similar to GP|23429518|gb APC2 {Arabidopsis thaliana},
partial (14%)
Length = 386
Score = 40.0 bits (92), Expect = 0.004
Identities = 24/96 (25%), Positives = 42/96 (43%)
Frame = +1
Query: 471 SWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELN 530
SWP NLP + L + + T R+L W+ ++G L+ F + ++ +
Sbjct: 4 SWPPIQDEPLNLPEPVDQLLSDYAKRFNEIKTPRKLQWKKSLGTIKLELQF-QDREIQFT 180
Query: 531 VSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKR 566
V+ ++M F + + K + A IPA L R
Sbjct: 181 VAPVHASIIMKFQDQPNWTSKNLAAAIGIPADVLNR 288
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,601,506
Number of Sequences: 63676
Number of extensions: 308528
Number of successful extensions: 1686
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1656
length of query: 703
length of database: 12,639,632
effective HSP length: 104
effective length of query: 599
effective length of database: 6,017,328
effective search space: 3604379472
effective search space used: 3604379472
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146568.7


