

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.5 - phase: 0
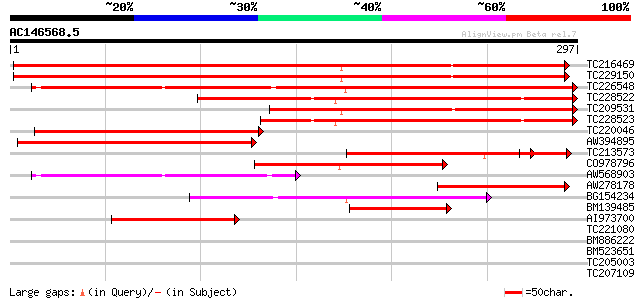
(297 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216469 UP|Q9SXM5 (Q9SXM5) Acidic chitinase, complete 382 e-107
TC229150 UP|Q9S7G9 (Q9S7G9) Acidic chitinase (Chitinase III-A) ,... 374 e-104
TC226548 UP|O48642 (O48642) Class III acidic endochitinase precu... 283 9e-77
TC228522 weakly similar to UP|Q6WDV9 (Q6WDV9) Chitinase class II... 220 5e-58
TC209531 similar to UP|O49876 (O49876) Class III chitinase precu... 200 6e-52
TC228523 similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 , ... 182 1e-46
TC220046 similar to UP|Q9XGB4 (Q9XGB4) Chitinase, partial (44%) 177 7e-45
AW394895 similar to SP|P29024|CHIA Acidic endochitinase precurso... 154 5e-38
TC213573 weakly similar to UP|Q9FUD7 (Q9FUD7) Class III acidic c... 117 8e-32
CO978796 128 3e-30
AW568903 similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class... 112 2e-25
AW278178 homologue to PIR|S57475|S574 chitinase (EC 3.2.1.14) cl... 109 1e-24
BG154234 weakly similar to PIR|T05187|T05 chitinase (EC 3.2.1.14... 88 5e-18
BM139485 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) clas... 85 4e-17
AI973700 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) clas... 78 4e-15
TC221080 similar to UP|Q8VQI8 (Q8VQI8) Chitinase (Fragment), par... 33 0.12
BM886222 weakly similar to PIR|S51591|S51 chitinase (EC 3.2.1.14... 31 0.59
BM523651 similar to SP|O04931|AGLU Alpha-glucosidase precursor (... 28 3.8
TC205003 similar to GB|AAS20952.1|42565013|AY531115 calmodulin b... 28 5.0
TC207109 similar to UP|Q9SZY4 (Q9SZY4) Nitrate transporter, part... 28 6.5
>TC216469 UP|Q9SXM5 (Q9SXM5) Acidic chitinase, complete
Length = 1113
Score = 382 bits (982), Expect = e-107
Identities = 189/294 (64%), Positives = 227/294 (76%), Gaps = 3/294 (1%)
Frame = +2
Query: 3 MELKSTISFTFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPT 62
M+ + S F LL L+L S+A IAIYWGQNG EGTLAEAC T NY+YV IAFL T
Sbjct: 26 MKTLNKASLILFPLLFLSLFKHSHAAGIAIYWGQNGGEGTLAEACNTRNYQYVNIAFLST 205
Query: 63 FGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANS 122
FG+GQTP +NLAGHCDP +NGCTGLSSDIK+CQ GIKVLLS+GGGAGSYS++S DA
Sbjct: 206 FGNGQTPQLNLAGHCDPNNNGCTGLSSDIKTCQDLGIKVLLSLGGGAGSYSLSSADDATQ 385
Query: 123 VATYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN--KKVYITAA 180
+A YLW NFLGG++ S PLG +LDGIDFDIE G S H+ DLAR L F+ +KVY++AA
Sbjct: 386 LANYLWQNFLGGQTGSGPLGNVILDGIDFDIESGGSDHYDDLARALNSFSSQRKVYLSAA 565
Query: 181 PQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFL 240
PQC PDA + A+ TGLFDYVWVQFYNNP CQY+ N ++W QW + +PA++IF+
Sbjct: 566 PQCIIPDAHLDRAIQTGLFDYVWVQFYNNPSCQYSSGNTNNLINSWNQWIT-VPASQIFM 742
Query: 241 GLPASPTAAGS-GFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSI 293
GLPAS AA S GF+ AD LTS VLPVIK SSKYGGVMLW+R++DVQ+GYS++I
Sbjct: 743 GLPASEAAAPSGGFVPADVLTSQVLPVIKQSSKYGGVMLWNRFNDVQNGYSNAI 904
>TC229150 UP|Q9S7G9 (Q9S7G9) Acidic chitinase (Chitinase III-A) , complete
Length = 1133
Score = 374 bits (960), Expect = e-104
Identities = 185/295 (62%), Positives = 224/295 (75%), Gaps = 4/295 (1%)
Frame = +2
Query: 3 MELKSTISFTFFSLLVLAL-ANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLP 61
M+ + S F LL L+L N S+A IA+YWGQNG EGTLAEAC TGNY+YV IAFL
Sbjct: 32 MKTPNKASLLLFPLLSLSLFINHSHAAGIAVYWGQNGGEGTLAEACNTGNYQYVNIAFLS 211
Query: 62 TFGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDAN 121
TFG+GQTP +NLAGHCDP +NGCTGLSSDI +CQ GIKVLLS+GGGAGSYS++S DA
Sbjct: 212 TFGNGQTPQLNLAGHCDPNNNGCTGLSSDINTCQDLGIKVLLSLGGGAGSYSLSSADDAT 391
Query: 122 SVATYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN--KKVYITA 179
+A YLW NFLGG++ S PLG +LDGIDFDIE G S H+ DLAR L F+ KVY++A
Sbjct: 392 QLANYLWENFLGGQTGSGPLGDVILDGIDFDIESGGSDHYDDLARALNSFSSQSKVYLSA 571
Query: 180 APQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIF 239
APQC PDA + A+ TGLFDYVWVQFYNNP CQY+ + ++W QW + +PA+ +F
Sbjct: 572 APQCIIPDAHLDAAIQTGLFDYVWVQFYNNPSCQYSSGNTNDLINSWNQWIT-VPASLVF 748
Query: 240 LGLPASPTAAGS-GFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSI 293
+GLPAS AA S GF+ AD LTS +LPVIK SS YGGVMLW R++DVQ+GYS++I
Sbjct: 749 MGLPASEAAAPSGGFVPADVLTSQILPVIKQSSNYGGVMLWDRFNDVQNGYSNAI 913
>TC226548 UP|O48642 (O48642) Class III acidic endochitinase precursor ,
complete
Length = 1274
Score = 283 bits (723), Expect = 9e-77
Identities = 150/292 (51%), Positives = 186/292 (63%), Gaps = 6/292 (2%)
Frame = +1
Query: 12 TFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMI 71
TFF + + S G I IYWGQN ++GTL C TGN+E V +AFL FG G TP
Sbjct: 82 TFF--FTIKPSQASTTGGITIYWGQNIDDGTLTSTCDTGNFEIVNLAFLNAFGCGITPSW 255
Query: 72 NLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNF 131
N AGHC + N C+ L I+ CQ KG+KV LS+GG G+YS+ S +DA VA YL+ NF
Sbjct: 256 NFAGHCGDW-NPCSILEPQIQYCQQKGVKVFLSLGGAKGTYSLCSPEDAKEVANYLYQNF 432
Query: 132 LGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFNKK----VYITAAPQCPFPD 187
L GK PLG L+GIDFDIE GS+ +WGDLA+ L + Y++AAPQC PD
Sbjct: 433 LSGKPG--PLGSVTLEGIDFDIELGSNLYWGDLAKELDALRHQNDHYFYLSAAPQCFMPD 606
Query: 188 AWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGI-PANKIFLGLPASP 246
+ NA+ TGLFD+V VQFYNNPPCQY+P ++W WTS + P N +F GLPASP
Sbjct: 607 YHLDNAIKTGLFDHVNVQFYNNPPCQYSPGNTQLLFNSWDDWTSNVLPNNSVFFGLPASP 786
Query: 247 TAAGS-GFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
AA S G+I L S VLP +K +S YGGVMLW RY DV + +S IK +V
Sbjct: 787 DAAPSGGYIPPQVLISEVLPYVKQASNYGGVMLWDRYHDVLNYHSDQIKDYV 942
>TC228522 weakly similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 ,
partial (57%)
Length = 725
Score = 220 bits (561), Expect = 5e-58
Identities = 108/203 (53%), Positives = 140/203 (68%), Gaps = 4/203 (1%)
Frame = +2
Query: 99 IKVLLSIGGGAGSYSIASTKDANSVATYLWNNFLGGKSSSRPLGPAVLDGIDFDIEGGSS 158
+KV+LSIGGG +YS++S DA VA Y+W NFLGGKS+SRP G A+LDG+DF+IE G
Sbjct: 8 VKVMLSIGGGTNTYSLSSPDDARQVADYIWANFLGGKSNSRPFGNAILDGVDFNIESGEL 187
Query: 159 QHWGDLARYLK----GFNKKVYITAAPQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQY 214
H+ LA L G KK Y+TA+PQC F + + ALTTGLFD+VW+QFYNNP C++
Sbjct: 188 -HYAALAYRLHDHYAGSKKKFYLTASPQCSFQNNLLHGALTTGLFDHVWIQFYNNPQCEF 364
Query: 215 NPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGSSKYG 274
F+ AW QWT+ I A K F+GLP+S AA +GF+S+ L + +LP+++ S KYG
Sbjct: 365 TSKDPTGFKSAWNQWTTSINAGKFFVGLPSSHAAARTGFVSSHALINNLLPIVR-SPKYG 541
Query: 275 GVMLWSRYDDVQSGYSSSIKSHV 297
GVMLW RY D+QS YS I V
Sbjct: 542 GVMLWDRYHDLQSRYSGKISGSV 610
>TC209531 similar to UP|O49876 (O49876) Class III chitinase precursor ,
partial (55%)
Length = 785
Score = 200 bits (509), Expect = 6e-52
Identities = 100/164 (60%), Positives = 119/164 (71%), Gaps = 3/164 (1%)
Frame = +3
Query: 137 SSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGFN--KKVYITAAPQCPFPDAWIGNAL 194
+SRP G AVLDGIDFDIE GS Q++GDLAR L ++ +KVY+ AAPQCP+PDA + +A+
Sbjct: 3 NSRPFGDAVLDGIDFDIEDGSGQYYGDLARELDAYSQQRKVYLAAAPQCPYPDAHLDSAI 182
Query: 195 TTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFLGLPASPTAAGS-GF 253
TGLFDYVWVQFYNNP C Y N AW QWTS A ++FLGLPAS AA S G+
Sbjct: 183 ATGLFDYVWVQFYNNPQCHYTSGNIDNLVSAWNQWTSS-EAKQVFLGLPASEAAAPSGGY 359
Query: 254 ISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
I D L S VLP I GS KYGGVM+W R++D QSGYS +IK+ V
Sbjct: 360 IPPDVLISDVLPAISGSPKYGGVMIWDRFNDGQSGYSDAIKASV 491
>TC228523 similar to UP|Q6WDV9 (Q6WDV9) Chitinase class III-1 , partial
(35%)
Length = 688
Score = 182 bits (463), Expect = 1e-46
Identities = 92/171 (53%), Positives = 118/171 (68%), Gaps = 5/171 (2%)
Frame = +2
Query: 132 LGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLK----GFNKKVYITAAPQCPFPD 187
LGGKS SRP G A+LDG+DFDIE G H+ LA L G +KK Y+TAAPQCPF +
Sbjct: 20 LGGKSKSRPFGDAILDGVDFDIESGEL-HYAALAYKLHDHYAGSSKKFYLTAAPQCPFQN 196
Query: 188 AWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGI-PANKIFLGLPASP 246
+ ALTTGLFD+VW++FY NP C++ P F+ AW QWT+ I A K F+GLPAS
Sbjct: 197 NLLHGALTTGLFDHVWIKFYTNPQCEFTPKDPTGFKSAWNQWTTSINAAGKFFVGLPASH 376
Query: 247 TAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDDVQSGYSSSIKSHV 297
AA +GF+S+ L + +LP+++ S KYGGVMLW R+ D+QSGYS I+ V
Sbjct: 377 AAARTGFVSSHALINHLLPIVR-SPKYGGVMLWDRFHDLQSGYSGKIRRSV 526
>TC220046 similar to UP|Q9XGB4 (Q9XGB4) Chitinase, partial (44%)
Length = 427
Score = 177 bits (448), Expect = 7e-45
Identities = 79/120 (65%), Positives = 100/120 (82%)
Frame = +2
Query: 14 FSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMINL 73
F LL+++L S+A IAIYWGQNGNEG+LA+AC TGNY++V IAFL TFG+GQTP +NL
Sbjct: 68 FPLLLISLFKSSHAAGIAIYWGQNGNEGSLADACNTGNYQFVNIAFLSTFGNGQTPQLNL 247
Query: 74 AGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNFLG 133
AGHC+P +NGCT S +IK+CQ KGIKVLLS+GG +GSYS+ S ++A +AT+LWNNFLG
Sbjct: 248 AGHCEPSTNGCTKFSDEIKACQGKGIKVLLSLGGASGSYSLGSAEEATQLATFLWNNFLG 427
>AW394895 similar to SP|P29024|CHIA Acidic endochitinase precursor (EC
3.2.1.14). [Adzuki bean Vigna angularis] {Phaseolus
angularis}, partial (42%)
Length = 425
Score = 154 bits (389), Expect = 5e-38
Identities = 73/126 (57%), Positives = 90/126 (70%), Gaps = 1/126 (0%)
Frame = +2
Query: 5 LKSTISFTFFSL-LVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTF 63
LK S F L L+ +L S+AG IA+YWGQNGNEG+L EAC T NY+YV I FL F
Sbjct: 47 LKQVSSLLLFPLFLIFSLFKSSHAGGIAVYWGQNGNEGSLEEACNTDNYQYVNIGFLNVF 226
Query: 64 GDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSV 123
G+ Q P +NLAGHC+P +N CTGLS+DI CQ+KGIKV LS+GG GSYS+ S +A +
Sbjct: 227 GNNQIPQLNLAGHCNPSTNECTGLSNDINVCQSKGIKVFLSLGGAVGSYSLNSASEATDL 406
Query: 124 ATYLWN 129
A YLW+
Sbjct: 407 AAYLWD 424
>TC213573 weakly similar to UP|Q9FUD7 (Q9FUD7) Class III acidic chitinase,
partial (31%)
Length = 504
Score = 117 bits (294), Expect(2) = 8e-32
Identities = 56/101 (55%), Positives = 68/101 (66%), Gaps = 2/101 (1%)
Frame = +3
Query: 177 ITAAPQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTSGIPAN 236
+TAAPQCPFPD AL+TGLFD+VWVQFYNN PCQ+ F+ +W QW S I A
Sbjct: 3 LTAAPQCPFPDEHQNGALSTGLFDFVWVQFYNNGPCQFESSDPTKFQKSWNQWISSIRAR 182
Query: 237 KIFLGLPASPT--AAGSGFISADDLTSTVLPVIKGSSKYGG 275
KI++GLPASP+ AGSGF+ L + VLP +K S K G
Sbjct: 183 KIYVGLPASPSPATAGSGFVPTRTLITQVLPFVKRSPKLWG 305
Score = 37.0 bits (84), Expect(2) = 8e-32
Identities = 16/27 (59%), Positives = 19/27 (70%)
Frame = +1
Query: 268 KGSSKYGGVMLWSRYDDVQSGYSSSIK 294
K YGGVMLW R D Q+GYS++IK
Sbjct: 283 KDRPSYGGVMLWDRAADKQTGYSTNIK 363
>CO978796
Length = 368
Score = 128 bits (322), Expect = 3e-30
Identities = 62/105 (59%), Positives = 72/105 (68%), Gaps = 4/105 (3%)
Frame = -2
Query: 129 NNFLGGKSSSRPLGPAVLDGIDFDIEGGSSQHWGDLARYLKGF----NKKVYITAAPQCP 184
N FLGGKSSSRP+G VLDGIDFDIE GS+Q + LAR K + NK+VYI AA QCP
Sbjct: 367 NTFLGGKSSSRPVGDXVLDGIDFDIELGSTQKYEHLARXXKAYSGVGNKRVYIGAARQCP 188
Query: 185 FPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQW 229
PD +G AL GLFD+VWVQ YNNPP QY+ +WK+W
Sbjct: 187 IPDRLLGTALNNGLFDFVWVQLYNNPPSQYDKGNINKLVSSWKRW 53
>AW568903 similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class III acidic
- soybean, partial (41%)
Length = 414
Score = 112 bits (280), Expect = 2e-25
Identities = 61/141 (43%), Positives = 76/141 (53%)
Frame = +3
Query: 12 TFFSLLVLALANGSNAGKIAIYWGQNGNEGTLAEACATGNYEYVIIAFLPTFGDGQTPMI 71
TFF + + G I IYWGQN + GTL C TGN+E V +AFL FG TP
Sbjct: 6 TFF--FTIKPSQAXTTGGITIYWGQNIDNGTLTSTCDTGNFEIVNLAFLNAFGCSITPSW 179
Query: 72 NLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNF 131
N GHC + N C+ L I+ CQ KG+K+ LS+ +YS+ S KD V YL+ NF
Sbjct: 180 NFTGHCGDW-NPCSILEPQIQYCQQKGVKIFLSLNSTKRTYSLYSPKDTKKVTNYLYQNF 356
Query: 132 LGGKSSSRPLGPAVLDGIDFD 152
L K PL L+ IDFD
Sbjct: 357 LSNK--PHPLRSITLESIDFD 413
>AW278178 homologue to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III
basic - cowpea, partial (24%)
Length = 380
Score = 109 bits (273), Expect = 1e-24
Identities = 51/69 (73%), Positives = 58/69 (83%)
Frame = -2
Query: 225 AWKQWTSGIPANKIFLGLPASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVMLWSRYDD 284
+W +WTS +PA KIFLGLPA P AAGSGFI AD LTS +LPVIK S KYGGVMLWSR+ D
Sbjct: 379 SWNRWTSTVPAGKIFLGLPADPAAAGSGFIPADTLTSEILPVIKESPKYGGVMLWSRFFD 200
Query: 285 VQSGYSSSI 293
VQ+GYS+SI
Sbjct: 199 VQNGYSTSI 173
>BG154234 weakly similar to PIR|T05187|T05 chitinase (EC 3.2.1.14) class III
acidic - soybean, partial (33%)
Length = 511
Score = 87.8 bits (216), Expect = 5e-18
Identities = 59/163 (36%), Positives = 83/163 (50%), Gaps = 5/163 (3%)
Frame = +2
Query: 95 QAKGIKVLLSIGGGAGSYSIASTKDANSVATYLWNNFLGGKSSSRPLGPAVLDGIDFDIE 154
Q +G+KV L++ G G+Y + S DA +A L++ L GK+S GI +I
Sbjct: 29 QERGVKVFLTLRGAKGTYYLCSLYDAQ*LACCLYHYLLMGKASL--F*SDS*KGIHINIA 202
Query: 155 GGSSQHWGDLARYLKGFNKKV----YITAAPQCPFPDAWIGNALTTGLFDYVWVQFYNNP 210
G + DL L ++ Y+ A QC D + +A+ TGL D+V +Q Y+
Sbjct: 203 LGIDIYG*DLVYELYAVPLQIDLYFYLCATAQCLMLDYHLYSAMKTGLSDHVNIQIYSTA 382
Query: 211 PCQYNPDAFMNFEDAWKQWTSGI-PANKIFLGLPASPTAAGSG 252
PCQY D +W WTS + P N +F GLPASP AA SG
Sbjct: 383 PCQYAADNTR*LCKSWDDWTSNVLPNNSVFFGLPASPDAAPSG 511
>BM139485 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III basic
- cowpea, partial (18%)
Length = 167
Score = 84.7 bits (208), Expect = 4e-17
Identities = 34/53 (64%), Positives = 39/53 (73%)
Frame = +3
Query: 179 AAPQCPFPDAWIGNALTTGLFDYVWVQFYNNPPCQYNPDAFMNFEDAWKQWTS 231
AAPQCP PD ++G AL TGLFD+VWVQFYNNPPCQY N +W +WTS
Sbjct: 9 AAPQCPIPDRFLGTALNTGLFDFVWVQFYNNPPCQYANGNITNLVSSWNRWTS 167
>AI973700 similar to PIR|S57475|S574 chitinase (EC 3.2.1.14) class III basic
- cowpea, partial (22%)
Length = 202
Score = 78.2 bits (191), Expect = 4e-15
Identities = 38/67 (56%), Positives = 50/67 (73%)
Frame = +2
Query: 54 YVIIAFLPTFGDGQTPMINLAGHCDPYSNGCTGLSSDIKSCQAKGIKVLLSIGGGAGSYS 113
++ I L T G+G+TP +NLA C+P +N CT SS+IK CQ+KGIKVLLSI GG GSY+
Sbjct: 2 HINIHSLNTLGNGKTPDMNLADDCNPTTNSCTKDSSEIKDCQSKGIKVLLSIRGGIGSYT 181
Query: 114 IASTKDA 120
+A +DA
Sbjct: 182 LALVEDA 202
>TC221080 similar to UP|Q8VQI8 (Q8VQI8) Chitinase (Fragment), partial (11%)
Length = 502
Score = 33.5 bits (75), Expect = 0.12
Identities = 30/119 (25%), Positives = 42/119 (35%), Gaps = 23/119 (19%)
Frame = +2
Query: 192 NALTTGLFDYVWVQFYNNPPCQYN-PDAFMNFEDAWKQW--TSGIPANKIFLGLP----- 243
N + LF PP +N P + E +W T G+P NK+ LGLP
Sbjct: 143 NVMAYDLFTSEGYPTVTQPPAPWNNPRGQFSAEQGVTEWNKTLGVPLNKLNLGLPFYGYK 322
Query: 244 -----------ASPTAAGSGFISADDLTSTVLPVIKGSSKYGGVML----WSRYDDVQS 287
+P G G + D+ + V+ S+ W YDD QS
Sbjct: 323 WSLSDSNKNGLFAPAKQGLGAVKYKDIKNVAAQVVFDSTYVTNYCFKGTDWFGYDDTQS 499
>BM886222 weakly similar to PIR|S51591|S51 chitinase (EC 3.2.1.14) / lysozyme
(EC 3.2.1.17) PZ precursor pathogenesis-related -
common, partial (32%)
Length = 425
Score = 31.2 bits (69), Expect = 0.59
Identities = 28/79 (35%), Positives = 38/79 (47%), Gaps = 21/79 (26%)
Frame = +1
Query: 76 HCDPYSNGCTGLSSD-------IKSCQAKG--IKVLLSIGGGAGSYSIAS----TKDANS 122
H DP +N T SSD ++ QAK +K LLSIGGG G A+ + AN+
Sbjct: 106 HLDPNTNKVTISSSDSSQFSTFTQTLQAKNPSVKTLLSIGGGFGPSLAANFSRMARQANT 285
Query: 123 VATYL--------WNNFLG 133
+++ NNFLG
Sbjct: 286 RKSFIDSSIQQARSNNFLG 342
>BM523651 similar to SP|O04931|AGLU Alpha-glucosidase precursor (EC 3.2.1.20)
(Maltase). [Sugar beet] {Beta vulgaris}, partial (14%)
Length = 437
Score = 28.5 bits (62), Expect = 3.8
Identities = 11/34 (32%), Positives = 20/34 (58%)
Frame = +1
Query: 207 YNNPPCQYNPDAFMNFEDAWKQWTSGIPANKIFL 240
+N P NP+ F+ F+D + Q +S +P+ + L
Sbjct: 10 FNTAPNPSNPETFLIFKDQYLQLSSSLPSQRASL 111
>TC205003 similar to GB|AAS20952.1|42565013|AY531115 calmodulin binding
protein 25 {Arabidopsis thaliana;} , partial (24%)
Length = 1133
Score = 28.1 bits (61), Expect = 5.0
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +1
Query: 232 GIPANKIFLGLPASPTAAGSG 252
G+PA+ +GLPA P+ AG G
Sbjct: 592 GVPADARHVGLPAGPSPAGGG 654
>TC207109 similar to UP|Q9SZY4 (Q9SZY4) Nitrate transporter, partial (61%)
Length = 1416
Score = 27.7 bits (60), Expect = 6.5
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 3/35 (8%)
Frame = -1
Query: 211 PCQYNPDAFMNFEDA---WKQWTSGIPANKIFLGL 242
P Y F+ F DA WK WT G+P ++L L
Sbjct: 210 PTLYGSCLFLFFIDAAITWKMWTMGLPKLLLYLYL 106
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,091,543
Number of Sequences: 63676
Number of extensions: 228996
Number of successful extensions: 927
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 902
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 903
length of query: 297
length of database: 12,639,632
effective HSP length: 97
effective length of query: 200
effective length of database: 6,463,060
effective search space: 1292612000
effective search space used: 1292612000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146568.5


