

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.2 - phase: 0
(494 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
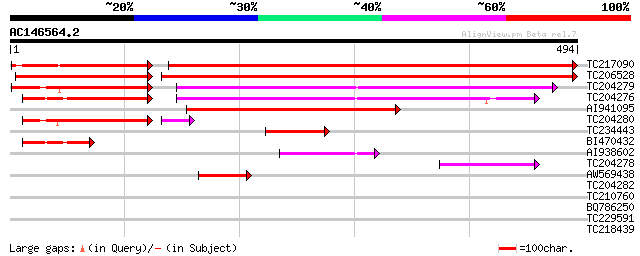
Score E
Sequences producing significant alignments: (bits) Value
TC217090 UP|VPE_SOYBN (P49045) Vacuolar processing enzyme precur... 432 e-169
TC206528 UP|Q9LLQ5 (Q9LLQ5) Seed maturation protein PM40, complete 416 e-166
TC204279 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase... 142 2e-63
TC204276 similar to UP|VPE1_PHAVU (O24325) Vacuolar processing e... 135 3e-60
AI941095 similar to SP|P49045|VPE_ Vacuolar processing enzyme pr... 184 8e-47
TC204280 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase... 139 2e-35
TC234443 similar to UP|VPE_SOYBN (P49045) Vacuolar processing en... 85 8e-17
BI470432 similar to GP|4589398|dbj| asparaginyl endopeptidase (V... 58 8e-09
AI938602 52 5e-07
TC204278 similar to UP|VPE1_PHAVU (O24325) Vacuolar processing e... 48 1e-05
AW569438 similar to GP|9622155|gb|A seed maturation protein PM40... 43 4e-04
TC204282 homologue to UP|Q9AUD9 (Q9AUD9) Asparaginyl endopeptida... 38 0.009
TC210760 weakly similar to UP|Q9EPR8 (Q9EPR8) Cdk3-binding prote... 31 1.4
BQ786250 30 1.9
TC229591 UP|O82585 (O82585) NADP-dependent isocitrate dehydrogen... 28 7.2
TC218439 similar to UP|Q9FXM3 (Q9FXM3) Replication factor C 37 k... 28 9.4
>TC217090 UP|VPE_SOYBN (P49045) Vacuolar processing enzyme precursor (VPE) ,
complete
Length = 1893
Score = 432 bits (1110), Expect(2) = e-169
Identities = 227/356 (63%), Positives = 260/356 (72%)
Frame = +2
Query: 139 DS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYR 198
D EP+ CYSWRQE* +G KWQ QQQ R +I+ILL WR W++WDAK+A+ +C+GFY
Sbjct: 614 DDGEPLCCYSWRQE*IEGRKWQSDQQQTRGQNIYILL*SWRSWNTWDAKHAIPLCHGFY* 793
Query: 199 RFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTC 258
EEETCIW LQ DG I SL*KW V*G +A GS LC NCI+CTRE*LGN+LSW G+
Sbjct: 794 CLEEETCIWKLQGDGYIRGSL*KWERV*GYNA*GSEYLCHNCIKCTRE*LGNLLSWNGSF 973
Query: 259 PTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*Y 318
TSRVHHLP GFV CLDGR * +QSKKG ETTIQ+G TDFK QQLC SCDAIW*Y
Sbjct: 974 STSRVHHLPRGFVQRCLDGR**GSQSKKGIRETTIQIGKATDFKFQQLCDGFSCDAIW*Y 1153
Query: 319 QHYR*KAIFVPRF*SCNGEPSSTQ*QIRI*NGSCKPKRCRDFIHVGNV*EIRSSNGKEKR 378
QH+ *KA+F+PRF*SC+ E ST+ Q R *NGSC PKRCR F+HV NV EI+ S K R
Sbjct: 1154QHHS*KALFIPRF*SCHCELPSTKRQARN*NGSC*PKRCRTFVHVANVSEIKPSVRK*DR 1333
Query: 379 NP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV 438
+P +CGD E *E+ RW* GIDWSFTVW+ KRFF ST R+ W C**L M KI GS V
Sbjct: 1334HPQTNCGDSEA*ETHRW*RGIDWSFTVWTRKRFFCSTIREGSWFVPC**LDMPKINGSGV 1513
Query: 439 *NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSMWWV*IRAITSIKQSL*C 494
*NSLW TD VW ETHASI +HLQQW F G +G G+ GS+ + AI SIKQ L C
Sbjct: 1514*NSLWDTDSVWHETHASIRQHLQQWRF*GLHGRGLFGSL*RLQCWAIASIKQRLQC 1681
Score = 182 bits (461), Expect(2) = e-169
Identities = 90/124 (72%), Positives = 104/124 (83%), Gaps = 1/124 (0%)
Frame = +3
Query: 2 FSIVLSLWSWLLLLLT-LDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS 60
+S+VL W++++L + G ARPN EWD VI+LP E VD A+ DEVGTRWAVLVAGS
Sbjct: 237 YSVVL----WMMVVLVRVHGAAARPNRKEWDSVIKLPTEPVD-ADSDEVGTRWAVLVAGS 401
Query: 61 SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV 120
+GYGNYRHQADVCHAYQLLIKGG+KEENIVVFMYDDIA NELNPR GVIINHP+G ++Y
Sbjct: 402 NGYGNYRHQADVCHAYQLLIKGGLKEENIVVFMYDDIATNELNPRHGVIINHPEGEDLYA 581
Query: 121 GVPK 124
GVPK
Sbjct: 582 GVPK 593
>TC206528 UP|Q9LLQ5 (Q9LLQ5) Seed maturation protein PM40, complete
Length = 1970
Score = 416 bits (1070), Expect(2) = e-166
Identities = 226/362 (62%), Positives = 259/362 (71%)
Frame = +1
Query: 133 LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCI 192
LH + D EP+ YSWRQE*S+G KWQ Q Q +I+ILL WR SSWDAK+A+ +
Sbjct: 472 LHR*ECDGPEPLCRYSWRQE*SEGRKWQSDQ*QT*GQNIYILL*SWRSGSSWDAKHAIPL 651
Query: 193 CYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNIL 252
CYG Y EEETCIW LQ+DG I SL*KW HV*G +A GS LC NCIQCTRE*LGN+L
Sbjct: 652 CYGLY*SLEEETCIWRLQEDGHIRGSL*KWEHV*GYNA*GSADLCHNCIQCTRE*LGNLL 831
Query: 253 SWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSC 312
SW G+ TSRVHHLP GFV CCLDGR * + SKKG ETTIQ+G TDFK QQLC SC
Sbjct: 832 SWNGSFSTSRVHHLPRGFVQCCLDGR**DS*SKKGVRETTIQIGKATDFKFQQLCDGFSC 1011
Query: 313 DAIW*YQHYR*KAIFVPRF*SCNGEPSSTQ*QIRI*NGSCKPKRCRDFIHVGNV*EIRSS 372
DAI *++H+ *KA+F+PRF*SC E ST+ + R *NGSC PKRCR F+HV NV EI+ S
Sbjct: 1012DAIR*HKHHS*KALFIPRF*SCRCELPSTERKARN*NGSC*PKRCRTFLHVANVSEIKPS 1191
Query: 373 NGKEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLK 432
KE R+P D GD E *E+ RW* GIDWSF VW+ KRFF ST + W C**L M K
Sbjct: 1192ARKEDRHPQTDSGDSEA*ETHRW*RGIDWSFIVWTRKRFFCSTIHEGSWSCPC**LDMPK 1371
Query: 433 IKGSVV*NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSMWWV*IRAITSIKQSL 492
I GS V*+SLW TD VW ETHASIC+HLQQ F G +G G+ GS+ + AITSIKQ L
Sbjct: 1372INGSGV*DSLWDTDSVWHETHASICQHLQQRCF*GLHGRGLCGSL*RLRFWAITSIKQRL 1551
Query: 493 *C 494
*C
Sbjct: 1552*C 1557
Score = 187 bits (476), Expect(2) = e-166
Identities = 90/120 (75%), Positives = 99/120 (82%), Gaps = 1/120 (0%)
Frame = +2
Query: 6 LSLWSWLLL-LLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYG 64
L LWSW+LL ++ G AR N EWD VI+LP E VD EVGTRWAVLVAGS+GYG
Sbjct: 110 LVLWSWMLLRMMMAQGAAARANRKEWDSVIKLPAEPVDADSDHEVGTRWAVLVAGSNGYG 289
Query: 65 NYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
NYRHQADVCHAYQLLIKGG+KEENIVVFMYDDIA +ELNPRPGVIINHP+G +VY GVPK
Sbjct: 290 NYRHQADVCHAYQLLIKGGLKEENIVVFMYDDIATDELNPRPGVIINHPEGQDVYAGVPK 469
>TC204279 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase (VmPE-1),
complete
Length = 1868
Score = 142 bits (358), Expect(2) = 2e-63
Identities = 76/129 (58%), Positives = 91/129 (69%), Gaps = 6/129 (4%)
Frame = +1
Query: 2 FSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVD------DAEVDEVGTRWAV 55
F+ ++ + L L+ TL L + H ++RLP E +A+ ++ GTRWAV
Sbjct: 88 FTPIMDRFPILFLVATLITLASGARH----DILRLPSEASRFFKAPANADQNDEGTRWAV 255
Query: 56 LVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQG 115
LVAGS+GY NYRHQ+DVCHAYQLL KGGVKEENIVVFMYDDIA NE NPRPGVIIN P G
Sbjct: 256 LVAGSNGYWNYRHQSDVCHAYQLLRKGGVKEENIVVFMYDDIAFNEENPRPGVIINSPHG 435
Query: 116 PNVYVGVPK 124
+VY GVPK
Sbjct: 436 NDVYKGVPK 462
Score = 119 bits (298), Expect(2) = 2e-63
Identities = 103/333 (30%), Positives = 163/333 (48%), Gaps = 1/333 (0%)
Frame = +3
Query: 146 CYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETC 205
CY+W+ S W+W+G + I+ILL WR S+ DA ++ +C EEE C
Sbjct: 504 CYTWK*VSSYWWQWEGCG*WPQ*SYIYILL*SWRSGSARDAY*SIHVCIRSD*SLEEEAC 683
Query: 206 IWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHH 265
W L K + +* W ++* ++ S LC+N +C R+ LGNILSWG + + + +
Sbjct: 684 FWNL*KPSILSRGM*IWEYL*RSSSRRSEYLCNNSFKCRRKQLGNILSWGVS*SSP*I*N 863
Query: 266 LPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*YQHYR*KA 325
LP V CCLDGRQ* Q N +TI++G D++ + + DA+W*++ R ++
Sbjct: 864 LPG*PVQCCLDGRQ*HTQFANRNFTSTIRIGQTKDYE-WKFNLWFPRDAVW*HRA*REQS 1040
Query: 326 IFVPRF*SCNGEPSSTQ*Q-IRI*NGSCKPKRCRDFIHVGNV*EIRSSNGKEKRNP*EDC 384
+ + SC *+ I S +P CR +G V + E + +
Sbjct: 1041RLIFGYKSC***FYFCA*KLIGATFKSSQPT*CRSHPFLG*VPQSSCGFF*ESCS*ETNS 1220
Query: 385 GDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV*NSLWV 444
+ ++R W +W K ++ + W T+C**LG+ G *++LW+
Sbjct: 1221*SNVSQNAYR*QHETYWKALLWH*KGSRTA*QC*TCWATTC**LGLP*NIG*DF*DTLWI 1400
Query: 445 TDPVWDETHASICKHLQQWYFRGFYGEGMHGSM 477
VWDET+ +CK LQ+W + G G+ SM
Sbjct: 1401PVSVWDETYEVLCKLLQRWNTKRANG*GLSTSM 1499
>TC204276 similar to UP|VPE1_PHAVU (O24325) Vacuolar processing enzyme
precursor (VPE) (Legumain-like proteinase) (LLP) ,
partial (98%)
Length = 1976
Score = 135 bits (339), Expect(2) = 3e-60
Identities = 72/113 (63%), Positives = 82/113 (71%)
Frame = +3
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQAD 71
LLLL+ V+ L D +RLP E +D GTRWAVL+AGS+GY NYRHQAD
Sbjct: 33 LLLLIAFATSVSGRRDLVGD-FLRLPSETDNDDNFK--GTRWAVLLAGSNGYWNYRHQAD 203
Query: 72 VCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
VCHAYQ+L KGG+KEENI+VFMYDDIA N NPRPGVIIN P G +VY GVPK
Sbjct: 204 VCHAYQILRKGGLKEENIIVFMYDDIAFNGENPRPGVIINKPDGGDVYKGVPK 362
Score = 115 bits (288), Expect(2) = 3e-60
Identities = 99/321 (30%), Positives = 156/321 (47%), Gaps = 5/321 (1%)
Frame = +2
Query: 146 CYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETC 205
C++W+ + W+W+G Q I IL PWR W + DA +++ I EE+ C
Sbjct: 404 CFTWK*VSTDWWQWEGCGQWS**SYICILY*PWRSWGARDACWSLLIRG*SD*SLEEKAC 583
Query: 206 IWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHH 265
W + K + +* W ++* + LC++C QC R+*LGNIL G + + R+++
Sbjct: 584 FWNI*KPSILSGGM*IWEYL*RSSS*RYQYLCNHCFQCRRK*LGNILPRGVS*SSPRIYN 763
Query: 266 LPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*YQHYR*KA 325
L V CCLDGRQ* Q N TI +G*R D+ + + + LS DA+W* + + +
Sbjct: 764 LFG*LVQCCLDGRQ*QTQFANRNSAPTI*IG*REDY-IWRFILWLSRDAVW*CRA*QRCS 940
Query: 326 IFVPRF*SCNGEPSSTQ*Q-IRI*NGSCKPKRCRDFIHVGNV*EIRSSNGKEKRNP*EDC 384
+ + + SC *+ + + + +P C +G V + +EK +
Sbjct: 941 LPLFGYRSC***FHFCG*KLLMVTFKTSQPT*C*SHPFLG*VPQSS*GFSQEKYSSETSF 1120
Query: 385 GDGET*ESFRW*CGIDWSFTVWSNKRFFSS----TGRKSDWPTSC**LGMLKIKGSVV*N 440
G + + R C DW +W K S+ TGR +C**L + G *+
Sbjct: 1121GSNVSQNACRQQCKTDWEAFIWH*KGSRSTQRC*TGRIG----TC**LALP*NHGEDF*D 1288
Query: 441 SLWVTDPVWDETHASICKHLQ 461
+LW+ VW ET+ +CKHLQ
Sbjct: 1289TLWILVSVWHETYEVLCKHLQ 1351
>AI941095 similar to SP|P49045|VPE_ Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE). [Soybean] {Glycine max}, partial (38%)
Length = 580
Score = 184 bits (467), Expect = 8e-47
Identities = 105/186 (56%), Positives = 125/186 (66%)
Frame = +1
Query: 155 KGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETCIWGLQKDGC 214
+G K Q QQQ R +I+ILL WR W++ DAK+A+ C+GFYR E+ETCIW +Q DG
Sbjct: 4 EGRKRQTDQQQTRGQNIYILL*SWRSWNTRDAKHAIPSCHGFYRCLEKETCIWKIQGDGY 183
Query: 215 IH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHHLPWGFV*CC 274
I SL*KW V*G +A GS L NCI+CTRE*LGN+L G+ TSRVHHLP FV C
Sbjct: 184 IPGSL*KWERV*GYNA*GSEYL*HNCIKCTRE*LGNLLLCNGSLYTSRVHHLPMRFVQSC 363
Query: 275 LDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*YQHYR*KAIFVPRF*SC 334
L GR + SKKG ETTI TDF LQ LC SCDA W YQ + *KAI +PRF*SC
Sbjct: 364 LYGR*RGSHSKKGIRETTIHTRYSTDFILQHLCHGFSCDATWRYQRHI*KAITIPRF*SC 543
Query: 335 NGEPSS 340
+ + +S
Sbjct: 544 HCDLTS 561
>TC204280 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase (VmPE-1),
partial (31%)
Length = 472
Score = 139 bits (351), Expect(2) = 2e-35
Identities = 73/118 (61%), Positives = 84/118 (70%), Gaps = 5/118 (4%)
Frame = +1
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEV-----VDDAEVDEVGTRWAVLVAGSSGYGNY 66
L LL TL L + H ++RLP E + ++ GTRWAVL+AGS+GY NY
Sbjct: 37 LFLLATLITLASGARH----DILRLPSEASTFFKAPGGDQNDEGTRWAVLIAGSNGYWNY 204
Query: 67 RHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
RHQ+DVCHAYQLL KGG+KEENIVVFMYDDIA NE NPRPGVIIN P G +VY GVPK
Sbjct: 205 RHQSDVCHAYQLLRKGGLKEENIVVFMYDDIAFNEENPRPGVIINSPHGNDVYKGVPK 378
Score = 27.7 bits (60), Expect(2) = 2e-35
Identities = 10/29 (34%), Positives = 17/29 (58%)
Frame = +3
Query: 133 LHWRQRDS*EPIRCYSWRQE*SKGWKWQG 161
LHW + + + CY+W+ S W+W+G
Sbjct: 381 LHW*RCNCWQLFCCYTWK*VSSYWWQWEG 467
>TC234443 similar to UP|VPE_SOYBN (P49045) Vacuolar processing enzyme
precursor (VPE) , partial (12%)
Length = 421
Score = 84.7 bits (208), Expect = 8e-17
Identities = 41/55 (74%), Positives = 43/55 (77%)
Frame = +3
Query: 224 HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHHLPWGFV*CCLDGR 278
HV*G A G LC NCIQCTRE*LGN+LSW G+ TSRVHHLP GFV CCLDGR
Sbjct: 78 HV*GYDA*GFEDLCHNCIQCTRE*LGNLLSWNGSFSTSRVHHLPRGFVQCCLDGR 242
>BI470432 similar to GP|4589398|dbj| asparaginyl endopeptidase (VmPE-1A)
{Vigna mungo}, partial (9%)
Length = 312
Score = 58.2 bits (139), Expect = 8e-09
Identities = 33/63 (52%), Positives = 40/63 (63%)
Frame = +2
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQAD 71
LLLL+ LV+ HL D +RLP E +D V GTRWAVL+AGS+GY NYRHQ
Sbjct: 20 LLLLIPFATLVSARPHLAGD-FLRLPSETDNDDNVQ--GTRWAVLLAGSNGYWNYRHQVH 190
Query: 72 VCH 74
+ H
Sbjct: 191LIH 199
>AI938602
Length = 325
Score = 52.4 bits (124), Expect = 5e-07
Identities = 33/87 (37%), Positives = 50/87 (56%)
Frame = +1
Query: 236 LCDNCIQCTRE*LGNILSWGGTCPTSRVHHLPWGFV*CCLDGRQ*VAQSKKGNGETTIQV 295
+C++C QC R+*L NIL G + + R+++L V CL+ Q* QS N TI +
Sbjct: 58 ICNHCFQCIRQ*LTNILPRGVS*ASPRIYNLTG*LVQ*CLNRIQ*PTQSANRNSAPTI*I 237
Query: 296 G*RTDFKLQQLCIRLSCDAIW*YQHYR 322
G*R D+ + + LS A+W* + Y+
Sbjct: 238 G*REDY-ISTFILWLSRHAVW*CKAYQ 315
>TC204278 similar to UP|VPE1_PHAVU (O24325) Vacuolar processing enzyme
precursor (VPE) (Legumain-like proteinase) (LLP) ,
partial (30%)
Length = 951
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/87 (32%), Positives = 43/87 (49%)
Frame = +2
Query: 375 KEKRNP*EDCGDGET*ESFRW*CGIDWSFTVWSNKRFFSSTGRKSDWPTSC**LGMLKIK 434
+EK + G + + R C DW +W K S+ + W +C**L + +
Sbjct: 83 QEKYSSETSFGSNVSQNACRQQCKTDWEAFIWH*KGSRSTQRC*TGWIGTC**LALPENH 262
Query: 435 GSVV*NSLWVTDPVWDETHASICKHLQ 461
G *++LW+ + DETH +CKHLQ
Sbjct: 263 GEDF*DTLWILVSIRDETHEVLCKHLQ 343
>AW569438 similar to GP|9622155|gb|A seed maturation protein PM40 {Glycine
max}, partial (8%)
Length = 154
Score = 42.7 bits (99), Expect = 4e-04
Identities = 21/46 (45%), Positives = 30/46 (64%)
Frame = +1
Query: 165 QIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETCIWGLQ 210
Q R +I+ILL WR +SWDAK+A+ +CY Y +E+T + LQ
Sbjct: 16 QTFRHNIYILL*SWRSGNSWDAKHAIPLCYVLY*TLDEKTYMSTLQ 153
>TC204282 homologue to UP|Q9AUD9 (Q9AUD9) Asparaginyl endopeptidase, partial
(12%)
Length = 606
Score = 38.1 bits (87), Expect = 0.009
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = +1
Query: 439 *NSLWVTDPVWDETHASICKHLQQWYFRGFYGEGMHGSM 477
*++LW++ VWDET+ +CK LQ+W G G+ SM
Sbjct: 172 *DTLWISVSVWDETYEVLCKLLQRWNTERANG*GLGTSM 288
>TC210760 weakly similar to UP|Q9EPR8 (Q9EPR8) Cdk3-binding protein ik3-1,
partial (4%)
Length = 826
Score = 30.8 bits (68), Expect = 1.4
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 8/54 (14%)
Frame = -2
Query: 154 SKGWKWQGHQQQIRR*DI------HILLRP--WRPWSSWDAKYAVCICYGFYRR 199
SKGW W +R+ + IL R W WS W A + IC GF RR
Sbjct: 318 SKGWIWTVGVGGVRKWKVLLEGSRWILERRIGWEFWSLWRAFWTTAICGGFERR 157
>BQ786250
Length = 409
Score = 30.4 bits (67), Expect = 1.9
Identities = 15/28 (53%), Positives = 22/28 (78%)
Frame = -1
Query: 1 MFSIVLSLWSWLLLLLTLDGLVARPNHL 28
+FS++L+LW WLLLL+ L L+ R NH+
Sbjct: 85 LFSLLLNLWVWLLLLV*L-LLLLRLNHI 5
>TC229591 UP|O82585 (O82585) NADP-dependent isocitrate dehydrogenase,
partial (97%)
Length = 1668
Score = 28.5 bits (62), Expect = 7.2
Identities = 12/27 (44%), Positives = 18/27 (66%)
Frame = +3
Query: 3 SIVLSLWSWLLLLLTLDGLVARPNHLE 29
S+ +WSW+L +LTL L+A P +E
Sbjct: 54 SLFSLIWSWILSILTLAFLIAMPLMIE 134
>TC218439 similar to UP|Q9FXM3 (Q9FXM3) Replication factor C 37 kDa subunit,
partial (50%)
Length = 961
Score = 28.1 bits (61), Expect = 9.4
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 8/51 (15%)
Frame = +2
Query: 181 WSSWDAKYAVCIC------YGFYRRFEEETCIWGLQKDGC--IH*SL*KW* 223
W S + + +C + F+R E + C+WG GC *SL KW*
Sbjct: 194 WRSSVGNHILAVCSSLIWIFNFFR--ESDFCVWGCSGKGCGGTS*SLQKW* 340
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.348 0.155 0.614
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,160,218
Number of Sequences: 63676
Number of extensions: 540009
Number of successful extensions: 4687
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 4610
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4677
length of query: 494
length of database: 12,639,632
effective HSP length: 101
effective length of query: 393
effective length of database: 6,208,356
effective search space: 2439883908
effective search space used: 2439883908
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146564.2


