

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.9 + phase: 0
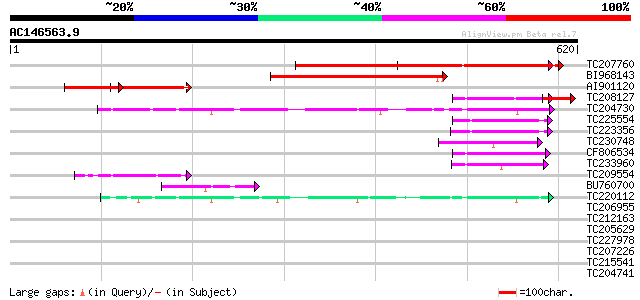
(620 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207760 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 495 e-140
BI968143 241 7e-64
AI901120 weakly similar to GP|15983787|gb| AT3g18370/MYF24_8 {Ar... 128 7e-30
TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 77 3e-14
TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, par... 70 2e-12
TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 59 6e-09
TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 54 2e-07
TC230748 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {A... 51 1e-06
CF806534 48 1e-05
TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%) 48 1e-05
TC209554 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {A... 46 6e-05
BU760700 44 2e-04
TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial ... 44 2e-04
TC206955 similar to GB|AAT41866.1|48310681|BT014883 At4g34150 {A... 41 0.001
TC212163 similar to UP|Q9LT26 (Q9LT26) Arabidopsis thaliana geno... 40 0.002
TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050),... 39 0.007
TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmi... 36 0.044
TC207226 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {A... 34 0.17
TC215541 similar to UP|Q84KK7 (Q84KK7) Respiratory burst oxidase... 32 1.1
TC204741 similar to UP|Q6PVY8 (Q6PVY8) Zinc finger protein, part... 30 2.4
>TC207760 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial
(40%)
Length = 1835
Score = 495 bits (1275), Expect = e-140
Identities = 241/294 (81%), Positives = 270/294 (90%), Gaps = 1/294 (0%)
Frame = +3
Query: 313 RRQQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTGT 372
RRQ +G++ G SED DD DL TFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDN GT
Sbjct: 9 RRQPNGTSXGCSEDNFDDXDLQTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNAGT 188
Query: 373 LRFNLYECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVV 432
LRFNLYE PNNV+CDYL SCEIKLRHVEDDSTIMWA+GPDSG+IAKQAQFCG+EIEMVV
Sbjct: 189 LRFNLYESCPNNVRCDYLASCEIKLRHVEDDSTIMWAIGPDSGVIAKQAQFCGEEIEMVV 368
Query: 433 PFEGTNSGELKVSIVVKEWQFSDGTHSLNNLR-NNSQQSLNGSSNIQLRTGKKLKITVVE 491
PFEG NSGELKVS+VVKEWQ+SDG+HSLN+LR ++SQQS+NGS N QLRTG+K+ +TVVE
Sbjct: 369 PFEGHNSGELKVSVVVKEWQYSDGSHSLNSLRSSSSQQSINGSPNFQLRTGRKINVTVVE 548
Query: 492 GKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFT 551
GKDL AAK+K+GKFDPYIKLQYGKV+QKT+T HTPNPVWNQ EFDE+GGGEYLKLK F+
Sbjct: 549 GKDL-AAKDKSGKFDPYIKLQYGKVVQKTRTVHTPNPVWNQMFEFDEIGGGEYLKLKGFS 725
Query: 552 EELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIKVDDQEGS 605
EE+FGDENIGSA VNLEGLV+GSVRDVWIPLERVRSGE+RL+I +I+ DDQEGS
Sbjct: 726 EEIFGDENIGSAHVNLEGLVEGSVRDVWIPLERVRSGELRLQI-SIRADDQEGS 884
Score = 80.1 bits (196), Expect = 3e-15
Identities = 55/171 (32%), Positives = 87/171 (50%), Gaps = 1/171 (0%)
Frame = +3
Query: 425 GDEIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKK 484
G ++ +P E SGEL++ I ++R + Q+ GS + L G
Sbjct: 789 GSVRDVWIPLERVRSGELRLQI---------------SIRADDQEGSKGSG-LGLGNGW- 917
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGE 543
+++ ++EG+DL AA + G DP++++ YG +KTK + T NP WNQT+EF + G
Sbjct: 918 IELVLIEGRDLVAADVR-GTSDPFVRVHYGNFKKKTKVIYKTLNPQWNQTLEFPDDGSQL 1094
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
L +K L +IG V + L D WIPL+ V+ GEI ++I
Sbjct: 1095MLYVKDHNA-LLPTSSIGECVVEYQRLPPNQTADKWIPLQGVKRGEIHIQI 1244
>BI968143
Length = 698
Score = 241 bits (615), Expect = 7e-64
Identities = 128/205 (62%), Positives = 151/205 (73%), Gaps = 12/205 (5%)
Frame = -2
Query: 286 RKKAVGGIIYVRVISANKLSSSSFKASR--RQQSGSTNGSSEDVSDDKDLHTFVEVEIEE 343
RK AVGGIIYV VISANKLS S FK+S RQQ+ + NG SE+ DD DL TFVEVE+EE
Sbjct: 697 RKTAVGGIIYVSVISANKLSRSCFKSSPSLRQQNSTINGYSENNLDDNDLQTFVEVEVEE 518
Query: 344 LTRRTDVRLGSTPRWDAPFNMVLHDNTGTLRFNLYECIPNNVKCDYLGSCEIKLRHVEDD 403
LTRRT + GS P WD FNMVLHDNTG +RFNLYEC + VKCD+L SCEIK+RHVEDD
Sbjct: 517 LTRRTGLSHGSNPMWDTTFNMVLHDNTGIVRFNLYECPSSGVKCDHLASCEIKMRHVEDD 338
Query: 404 STIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVS--IVVKEWQFSDGTHSLN 461
STIMWA+GPDS IAK A+FCGDE+EMVVPFEGTNS ELKV +V+ + +F H L
Sbjct: 337 STIMWAIGPDSSAIAKHAKFCGDEVEMVVPFEGTNSVELKVKFVVVLLKVEFDPHEHHLP 158
Query: 462 NLRNN-----SQQSL---NGSSNIQ 478
N+ + Q++ NG+ N+Q
Sbjct: 157 NVIRS*YFDFPMQNIAHGNGNHNVQ 83
>AI901120 weakly similar to GP|15983787|gb| AT3g18370/MYF24_8 {Arabidopsis
thaliana}, partial (8%)
Length = 411
Score = 128 bits (322), Expect = 7e-30
Identities = 64/89 (71%), Positives = 72/89 (79%)
Frame = +2
Query: 111 PNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRV 170
P PK S RL AIVE RLKLRKPRF+E+VE+QEFSLGSCPPSL LQGMRWST G QRV
Sbjct: 152 PTILTPKFSRRLKAIVEKRLKLRKPRFIEKVEVQEFSLGSCPPSLGLQGMRWSTSGGQRV 331
Query: 171 MQLGFDWDTHEMSILLLAKLAKPLMGTAR 199
++ FDWDT EMSIL+LAKL+ +GTAR
Sbjct: 332 LKTSFDWDTSEMSILMLAKLS---VGTAR 409
Score = 107 bits (266), Expect = 2e-23
Identities = 44/64 (68%), Positives = 56/64 (86%)
Frame = +1
Query: 61 VWATIQYGRYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSS 120
+WA++QYG YQRKLL E+L+KKWKRI+LN SP+TPLEHCEWLN LLT+IW NYFNPK+
Sbjct: 1 LWASMQYGNYQRKLLEEELNKKWKRILLNTSPMTPLEHCEWLNLLLTQIWSNYFNPKVLK 180
Query: 121 RLSA 124
++ +
Sbjct: 181 KVKS 192
>TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial
(23%)
Length = 845
Score = 76.6 bits (187), Expect = 3e-14
Identities = 43/111 (38%), Positives = 65/111 (57%), Gaps = 1/111 (0%)
Frame = +3
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGE 543
+++ V+E +DL AA + G DPY+++ YG ++TK H T NP WNQT+EF + G
Sbjct: 258 IELVVIEARDLIAA-DLRGTSDPYVRVNYGNSKKRTKVIHKTLNPRWNQTLEFLDDGSPL 434
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
L +K L + +IG V + L + D WIPL+ V+SGEI ++I
Sbjct: 435 ILHVKDHNA-LLPESSIGEGVVEYQRLPPNQMSDKWIPLQGVKSGEIHIQI 584
Score = 53.9 bits (128), Expect = 2e-07
Identities = 25/36 (69%), Positives = 31/36 (85%)
Frame = +1
Query: 583 ERVRSGEIRLKIEAIKVDDQEGSTVCFFSQKLSILL 618
E V SGE++LKIE +KV+DQEGS VCFF++KL ILL
Sbjct: 1 EGVSSGELKLKIEVVKVEDQEGSRVCFFNRKLVILL 108
>TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, partial (97%)
Length = 2084
Score = 70.5 bits (171), Expect = 2e-12
Identities = 105/522 (20%), Positives = 219/522 (41%), Gaps = 23/522 (4%)
Frame = +1
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNK + +WP Y + + +I + + + P++ ++ VE +E +LGS PP+
Sbjct: 406 DRLDWLNKFILYMWP-YLDKAICKTARSIAKPIIAEQIPKYKIDSVEFEELNLGSLPPT- 579
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ + T + +M+ W + +++A A L T ++V + +
Sbjct: 580 -FQGMKVYVTDEKELIMEPSVKWAGNPN--IIVAIKAFGLRATVQVVDLQVFAAPRITLK 750
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + S + P V G+ + S+ PG+ ++++ D + K
Sbjct: 751 PLVPSFPCFANIYVSLMEKPHVDFGLKLLGADAMSI-----PGLYRIVQEIIKDQVAKMY 915
Query: 271 VEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVSDD 330
+ P+ + K GI++V+V+ A KL +D+
Sbjct: 916 LWPKALEVQIMDPTKAMKVPVGILHVKVVRAEKL------------------KKKDLLGA 1041
Query: 331 KDLHTFVEVEIEEL-TRRTDVRLGS-TPRWDAPFNMVLHD-NTGTLRFNLYECIPNNVKC 387
D + +++ E+L +++T V+ + P W+ FN+V+ D + L +Y+ K
Sbjct: 1042SDPYVKLKLTEEKLPSKKTTVKYKNLNPEWNEEFNIVVKDPESQVLELTVYDW-EQIGKH 1218
Query: 388 DYLGSCEIKLRHVEDDS------TIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGE 441
D +G I L+ + D ++ + P+ P + G+
Sbjct: 1219DKMGMNVIPLKEITPDEPKAVTLNLLKTMDPND------------------PENAKSRGQ 1344
Query: 442 LKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEK 501
L V ++ K ++ + S + N +++ G+ +G L I V E +D+ E
Sbjct: 1345LTVEVLYKPFKEDELPQSAED-SNAIEKAPEGTP----ASGGLLVIIVHEAEDV----EG 1497
Query: 502 TGKFDPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE----- 553
+PY++L + +KTK +P W ++ +F +E E L ++V +
Sbjct: 1498KHHTNPYVRLLFKGEERKTKHVKKNRDPRWGESFQFMLEEPPTNERLYVEVQSASSKLGL 1677
Query: 554 LFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
L E++G + L +V + L R+G I+++++
Sbjct: 1678LHPKESLGYVDIKLSDVVTNKRINEKYHLIDSRNGRIQIELQ 1803
>TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (95%)
Length = 1383
Score = 58.9 bits (141), Expect = 6e-09
Identities = 42/112 (37%), Positives = 63/112 (55%), Gaps = 3/112 (2%)
Frame = +3
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP-NPVWNQTIEFDEVGGGE 543
LK+ VV+GK L KT DPY+ L+ G KTK ++ NPVWN+ + F
Sbjct: 306 LKVIVVQGKRLVIRDFKTS--DPYVVLKLGNQTAKTKVINSCLNPVWNEELNFTLTEPLG 479
Query: 544 YLKLKVFTEELF-GDENIGSAQVNLEGLVDGS-VRDVWIPLERVRSGEIRLK 593
L L+VF ++L D+ +G+A +NL+ +V + +RD+ RV SGE L+
Sbjct: 480 VLNLEVFDKDLLKADDKMGNAFLNLQPIVSAARLRDIL----RVSSGETTLR 623
>TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (96%)
Length = 705
Score = 53.9 bits (128), Expect = 2e-07
Identities = 38/114 (33%), Positives = 63/114 (54%), Gaps = 3/114 (2%)
Frame = +3
Query: 483 KKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP-NPVWNQTIEFDEVGG 541
K LK+ VV+GK L K+ DPY+ ++ G KT+ NPVWN+ + F
Sbjct: 81 KILKVIVVQGKRLVIRDFKSS--DPYVVVKLGNQTAKTRVIRCCLNPVWNEELNFTLTEP 254
Query: 542 GEYLKLKVFTEELF-GDENIGSAQVNLEGLVDGS-VRDVWIPLERVRSGEIRLK 593
L L+VF ++L+ D+ +G++ +NL+ L+ + +RD+ +V SGE L+
Sbjct: 255 LGVLNLEVFDKDLWKADDKMGNSYLNLQPLISAARLRDIL----KVSSGETTLR 404
>TC230748 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {Arabidopsis
thaliana;} , partial (48%)
Length = 903
Score = 51.2 bits (121), Expect = 1e-06
Identities = 38/119 (31%), Positives = 59/119 (48%), Gaps = 6/119 (5%)
Frame = +1
Query: 470 SLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPN-- 527
+L GS ++ R+ L + K+ GK DPY + + ++ K S T N
Sbjct: 205 TLPGSFSLPARS*LGLTRISPRSGPMLTNKDIIGKSDPYAVVYIRPLRERMKKSKTINND 384
Query: 528 --PVWNQTIEF-DEVGGGEYLKLKVF-TEELFGDENIGSAQVNLEGLVDGSVRDVWIPL 582
P+WN+ EF E +++ +KV+ +E L E IG AQ+ L L G V+DVW+ L
Sbjct: 385 LNPIWNEHFEFVVEDVSTQHVTVKVYDSEGLQSSELIGCAQLQLSELQPGKVKDVWLKL 561
>CF806534
Length = 512
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/113 (26%), Positives = 62/113 (54%), Gaps = 6/113 (5%)
Frame = +3
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS--HTPNPVWNQTIEF--DEVG 540
+++ +V+ K L A + G+ DPY+ +QY Q++ + NPVWN+ F + +G
Sbjct: 90 MEVQLVKAKGLHNA-DIFGEMDPYVLIQYNDQEQRSSVAIGQGTNPVWNEKFMFKVEYLG 266
Query: 541 GGEYLKL--KVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIR 591
G+ KL K+ ++L+ DE +G A ++++ L+ + + L+ ++ +R
Sbjct: 267 SGDKHKLIFKIMDQDLYTDEFVGQATIHVKDLLAQGIDNGGAKLQTLKYRVVR 425
>TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%)
Length = 862
Score = 47.8 bits (112), Expect = 1e-05
Identities = 31/110 (28%), Positives = 58/110 (52%), Gaps = 4/110 (3%)
Frame = +2
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTP-NPVWNQTIEF---DEV 539
KLKI V+ L AA ++ G DP++ + G + KT + NPVWN+ ++F ++
Sbjct: 86 KLKIRVISASSLQAA-DRGGTSDPFVHFKQGHMGVKTTVKNKELNPVWNEDLQFGIYEKD 262
Query: 540 GGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGE 589
L ++V +++ D+ IG+ +V+L + R+ + L +SG+
Sbjct: 263 AQTGTLSIEVLDKDMLSDDCIGTYKVSLSEVPHNQPREYDVHLTGGQSGK 412
>TC209554 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {Arabidopsis
thaliana;} , partial (32%)
Length = 614
Score = 45.8 bits (107), Expect = 6e-05
Identities = 38/127 (29%), Positives = 58/127 (44%)
Frame = +1
Query: 72 RKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLK 131
RK+L W ++ ++S + WLN LT+IWP Y N S + A VE L+
Sbjct: 208 RKILPPQFYPSW--VVFSSS-----QKLTWLNSHLTKIWP-YVNEAASELIKASVEPILE 363
Query: 132 LRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLA 191
+P L ++ +F+LG+ P + GD M+L WD + IL +
Sbjct: 364 EYRPVVLAALKFSKFTLGTVAPQFTGVSI-IEDGGDGVTMELEMQWDGNPSIILDI---- 528
Query: 192 KPLMGTA 198
K L+G A
Sbjct: 529 KTLLGVA 549
>BU760700
Length = 445
Score = 43.9 bits (102), Expect = 2e-04
Identities = 40/118 (33%), Positives = 62/118 (51%), Gaps = 11/118 (9%)
Frame = +3
Query: 167 DQRVMQLGFDWDT-HEMSILLLAKLAKPL-MGT-ARIVINSLHIKGDLI----FTPILD- 218
D V++LG ++ T +MS +L KL K L G A++ I +H++G ++ F P
Sbjct: 45 DHLVLELGMNFLTADDMSAILAVKLRKRLGFGMWAKLHITGMHVEGKVLVGVKFLPTWPF 224
Query: 219 -GKALLYSFVSAP--EVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEP 273
G+ L FV P ++ V F G L TE PG++ WL+KL + +T+VEP
Sbjct: 225 IGR-LRVCFVEPPYFQMTVKPMFTHG----LDVTELPGIAGWLDKLLSIAFEQTLVEP 383
>TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial (76%)
Length = 1867
Score = 43.9 bits (102), Expect = 2e-04
Identities = 110/535 (20%), Positives = 206/535 (37%), Gaps = 40/535 (7%)
Frame = +2
Query: 100 EWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLE--------RVELQEFSLGSC 151
+WLNK L + W P L + + I+ +R +P F E +E + SLG+
Sbjct: 311 DWLNKFLLDTW-----PFLDTAICKIIRSR---AQPIFFEYIGKYQIKAIEFDKLSLGTL 466
Query: 152 PPSLALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGD 210
PP++ G++ T G + VM+ W + ++L+ L T ++V +
Sbjct: 467 PPTVC--GIKVLETNGKELVMEQVIKWAGNPE--IVLSVYVASLKITVQLVDLQIFAAPR 634
Query: 211 LIFTPILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTL 266
+ P++ ++ S + P V G+ G S+ PG+ ++++ +
Sbjct: 635 VTLRPLVPTFPCFANIVVSLMEKPHVDFGMNVSGGDIMSI-----PGLYRFVQETIKKQV 799
Query: 267 VKTMVEPRRRCFTLPAVDLRKKAVG---GIIYVRVISANKLSSSSFKASRRQQSGSTNGS 323
+ P + +P +D A+ GI++V V+ A KL
Sbjct: 800 ANLYLWP--QTLEIPILDESTVAIKKPVGILHVNVVRAQKLLKM---------------- 925
Query: 324 SEDVSDDKDLHTFVEVEIEEL-TRRTDV-RLGSTPRWDAPFNMVLHD-NTGTLRFNLY-- 378
D+ D + + + ++L ++T V R P W+ F +V+ D + L+ +Y
Sbjct: 926 --DLLGTSDPYVKLSLTGDKLPAKKTTVKRKNLNPEWNEKFKIVVKDPQSQVLQLQVYDW 1099
Query: 379 -----------ECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDE 427
+ +P V Y E L ++D + G I F
Sbjct: 1100DKVGGHDKLGMQLVPLKVLKPYENK-EFTLDLLKDTNLNETPHKKPRGKIVVDLTF---- 1264
Query: 428 IEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKI 487
VPF KE G S R S + +Q G L I
Sbjct: 1265----VPF--------------KEDSNKFGGPSEGYSRKESGIDIVSDDEVQEGAG-LLSI 1387
Query: 488 TVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT-SHTPNPVWNQTIEF--DEVGGGEY 544
+ E +++ E +P+ L + ++TKT T +P WN+ +F +E E
Sbjct: 1388VIQEAEEV----EGDHHNNPFAVLTFRGEKKRTKTMKKTRHPRWNEEFQFMLEEPPLHEK 1555
Query: 545 LKLKVFTE----ELFGDENIGSAQVNLEGLV-DGSVRDVWIPLERVRSGEIRLKI 594
+ ++V ++ E++G ++NL +V +G + D + L R+G + ++I
Sbjct: 1556IHIEVMSKRKNFSFLPKESLGHVEINLRDVVHNGRINDKY-HLINSRNGVMHVEI 1717
>TC206955 similar to GB|AAT41866.1|48310681|BT014883 At4g34150 {Arabidopsis
thaliana;} , partial (52%)
Length = 498
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/139 (27%), Positives = 67/139 (47%), Gaps = 6/139 (4%)
Frame = +2
Query: 466 NSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYG--KVMQKTKTS 523
+S S++ + IQ G+ L++TVV L E + DPY+ ++YG K +T T
Sbjct: 71 SSSDSMSSITGIQ---GQPLEVTVVSCSKLKDT-EWISRQDPYVCVEYGSTKFRTRTCTD 238
Query: 524 HTPNPVWNQTIEFDEVGGGEYLKLKVF-TEELFGDENIGSAQVNLEGLVDGSVRDVWIPL 582
NPV+ + F + G L + V+ + L D+ IGS ++ L ++ D PL
Sbjct: 239 GGKNPVFQEKFIFPLIEGLRELNVLVWNSNTLTFDDFIGSGKIQLHKVLSQGFDDSAWPL 418
Query: 583 ERVR---SGEIRLKIEAIK 598
+ +GE+++ + K
Sbjct: 419 QTKTGRYAGEVKVILHYAK 475
>TC212163 similar to UP|Q9LT26 (Q9LT26) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MPN9, partial (30%)
Length = 739
Score = 40.4 bits (93), Expect = 0.002
Identities = 25/92 (27%), Positives = 46/92 (49%)
Frame = +3
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLA 156
E EW+N +L ++W Y + + + L+KP +++RVE+++FSLG P S+
Sbjct: 369 ESVEWVNMVLGKLWKVYRGGIENWIIGLLQPVIDNLKKPDYVQRVEIKQFSLGDEPLSVR 548
Query: 157 LQGMRWSTIGDQRVMQLGFDWDTHEMSILLLA 188
R S + Q+G + +L+L+
Sbjct: 549 NVERRTSRRVNDLQYQIGLRYTGGARMLLMLS 644
>TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050), partial
(92%)
Length = 1766
Score = 38.9 bits (89), Expect = 0.007
Identities = 31/112 (27%), Positives = 56/112 (49%), Gaps = 1/112 (0%)
Frame = +3
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLA 156
E +WLNK LT++WP + + + VE L+ +P + ++ + SLG+ P
Sbjct: 108 EQVKWLNKKLTKLWP-FVAEAATMVIRESVEPLLEEYRPPGITSLKFSKLSLGNVAPK-- 278
Query: 157 LQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHI 207
++G+R S Q +M + F W + SI+L + A L+ + I + L +
Sbjct: 279 IEGIRVQSLTKGQIIMDIDFRWG-GDPSIILAVEAA--LVASIPIQLKDLQV 425
>TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmin
{Arabidopsis thaliana;} , partial (72%)
Length = 1470
Score = 36.2 bits (82), Expect = 0.044
Identities = 78/390 (20%), Positives = 160/390 (41%), Gaps = 19/390 (4%)
Frame = +2
Query: 225 SFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCFTLPAVD 284
S + P V G+ S+P GV +++L D + + P+ + +D
Sbjct: 191 SLMEKPHVDFGLKLIGADLMSIP-----GVYRIVQELIKDQVANMYLWPKT--LEVQVLD 349
Query: 285 LRK--KAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVSDDKDLHTFVEVEIE 342
+ K K GI++V+V+ A KL G+S+ +V++++
Sbjct: 350 MSKALKRPVGILHVKVLQAMKLKKKDLL-----------GASDP---------YVKLKLT 469
Query: 343 E---LTRRTDVRLGS-TPRWDAPFNMVLHD-NTGTLRFNLYECIPNNVKCDYLGSCEIKL 397
E +++T V+ + P+W+ FN+V+ D ++ + N+Y+ K D +G I L
Sbjct: 470 EDKWPSKKTTVKHKNLNPQWNEEFNIVVKDPDSQVIEINVYDW-EQVGKHDKMGMNVIPL 646
Query: 398 RHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNS---GELKVSIVVKEWQFS 454
+ V + T +F D ++ + P + N G++ V + K ++
Sbjct: 647 KEVSPEET---------------KRFTLDLLKNMDPNDAQNEKSRGQIVVELTYKPFKEE 781
Query: 455 DGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKF--DPYIKLQ 512
D Q++ + G L + V E +D+ GK+ +P+++L
Sbjct: 782 DLGKGF-----EETQTVPKAPEGTPAGGGLLVVIVHEAQDV------EGKYHTNPHVRLI 928
Query: 513 YGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE----LFGDENIGSAQV 565
+ +KTK +P W +F DE + L ++V + L E++G +
Sbjct: 929 FRGDEKKTKRIKKNRDPRWEDEFQFMVDEPPTNDRLHVEVVSTSSRNLLHQKESLGYIDI 1108
Query: 566 NLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
NL +V + L ++G ++++++
Sbjct: 1109NLGDVVANKRINEKYHLIDSKNGRLQIELQ 1198
>TC207226 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {Arabidopsis
thaliana;} , partial (16%)
Length = 621
Score = 34.3 bits (77), Expect = 0.17
Identities = 21/71 (29%), Positives = 42/71 (58%), Gaps = 2/71 (2%)
Frame = +1
Query: 527 NPVWNQTIEF-DEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLV-DGSVRDVWIPLER 584
NPVWNQT +F E G + L ++V+ + FG + +G + L ++ +G ++ ++ L+
Sbjct: 43 NPVWNQTFDFVVEDGLHDMLIVEVWDHDTFGKDYMGRCILTLTRVILEGEYKERFV-LDG 219
Query: 585 VRSGEIRLKIE 595
+SG + L ++
Sbjct: 220 AKSGFLNLHLK 252
>TC215541 similar to UP|Q84KK7 (Q84KK7) Respiratory burst oxidase homolog,
partial (54%)
Length = 1695
Score = 31.6 bits (70), Expect = 1.1
Identities = 15/42 (35%), Positives = 23/42 (54%)
Frame = -2
Query: 444 VSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKL 485
VS + Q+SDG ++ + N S Q +NG N Q+ G+ L
Sbjct: 680 VSRIKSHLQYSDGLNARSECSNQSFQCINGDGNSQIHPGRFL 555
>TC204741 similar to UP|Q6PVY8 (Q6PVY8) Zinc finger protein, partial (80%)
Length = 1590
Score = 30.4 bits (67), Expect = 2.4
Identities = 14/37 (37%), Positives = 24/37 (64%)
Frame = +1
Query: 372 TLRFNLYECIPNNVKCDYLGSCEIKLRHVEDDSTIMW 408
TLR L+EC+ +N+K + GSC + + + S+I+W
Sbjct: 1042 TLRLLLHECL-SNIKIKWFGSCAMTVVKLLTFSSILW 1149
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,876,437
Number of Sequences: 63676
Number of extensions: 396401
Number of successful extensions: 1771
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 1750
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1760
length of query: 620
length of database: 12,639,632
effective HSP length: 103
effective length of query: 517
effective length of database: 6,081,004
effective search space: 3143879068
effective search space used: 3143879068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146563.9


