

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.12 - phase: 0
(336 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
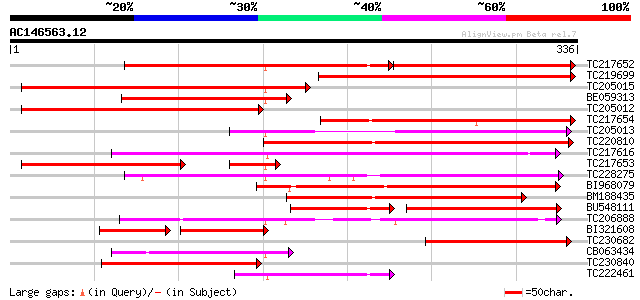
Score E
Sequences producing significant alignments: (bits) Value
TC217652 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, part... 218 1e-96
TC219699 287 4e-78
TC205015 212 2e-55
BE059313 189 2e-48
TC205012 similar to UP|Q8CD59 (Q8CD59) Mus musculus 13 days embr... 182 1e-46
TC217654 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, part... 174 5e-44
TC205013 weakly similar to UP|Q9LF16 (Q9LF16) Lipase-like protei... 164 5e-41
TC220810 similar to PIR|T00552|T00552 lysophospholipase homolog ... 160 6e-40
TC217616 similar to GB|AAM63479.1|21554372|AY086477 phospholipas... 159 2e-39
TC217653 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, part... 130 3e-38
TC228275 weakly similar to UP|Q9M3D0 (Q9M3D0) Lipase-like protei... 153 9e-38
BI968079 weakly similar to PIR|H86244|H86 lysophospholipase homo... 144 6e-35
BM188435 140 1e-33
BU548111 106 3e-33
TC206888 similar to UP|Q8LEE3 (Q8LEE3) Lysophospholipase-like pr... 118 3e-27
BI321608 82 4e-26
TC230682 101 5e-22
CB063434 96 3e-20
TC230840 weakly similar to PIR|T00552|T00552 lysophospholipase h... 90 2e-18
TC222461 similar to UP|Q8RZT4 (Q8RZT4) Phospholipase-like protei... 82 3e-16
>TC217652 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, partial (71%)
Length = 1204
Score = 218 bits (554), Expect(2) = 1e-96
Identities = 104/172 (60%), Positives = 129/172 (74%), Gaps = 13/172 (7%)
Frame = +1
Query: 69 KVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGL 128
++F KSW+P S K V+YCHGY DTC+F+FEG+ARKLASSG+ VFA+DYPGFGLS+GL
Sbjct: 1 EIFCKSWLPSASKPKAAVFYCHGYGDTCSFFFEGIARKLASSGYAVFAMDYPGFGLSEGL 180
Query: 129 HGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAAL 175
H YI SF+ LV+DVIEH+SKIK G+SMGGA+AL IH KQP AWDGA L
Sbjct: 181 HCYIHSFDGLVDDVIEHYSKIKENPEFHSLPSFLFGQSMGGAVALKIHLKQPKAWDGAIL 360
Query: 176 IAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRE 227
+AP+CK A+DM+P + ILIG+A VLPK KLVP K ++ E +RD +KRE
Sbjct: 361 VAPMCKIADDMVPPKFLTHILIGLANVLPKHKLVPNK-DLAEAAFRDLKKRE 513
Score = 153 bits (387), Expect(2) = 1e-96
Identities = 72/108 (66%), Positives = 87/108 (79%)
Frame = +3
Query: 228 LAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQ 287
L YNV+ YKDKPRL +A+E+LK T+E+EQRL+EVSLP+ ++HGEAD +TDPS SKALY+
Sbjct: 591 LTAYNVVAYKDKPRLKSAVEMLKTTEEIEQRLKEVSLPIFILHGEADTVTDPSVSKALYE 770
Query: 288 KAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
A DKKL LYKDA+H LLEGEPDE I V DIISWLD+HS T N+
Sbjct: 771 NASCSDKKLQLYKDAYHALLEGEPDEIITQVFGDIISWLDEHSLTHNQ 914
>TC219699
Length = 696
Score = 287 bits (735), Expect = 4e-78
Identities = 141/152 (92%), Positives = 147/152 (95%)
Frame = +2
Query: 184 EDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLG 243
EDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVK+NI+RD KR+LAPYNVL YKDKPRLG
Sbjct: 2 EDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKDNIFRDVNKRKLAPYNVLLYKDKPRLG 181
Query: 244 TALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAF 303
TALELLKATQELEQRLEEVSLPLL+MHGEADIITDPSASKALY+KAKVKDKKLCLYKDAF
Sbjct: 182 TALELLKATQELEQRLEEVSLPLLIMHGEADIITDPSASKALYEKAKVKDKKLCLYKDAF 361
Query: 304 HTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
HTLLEGEPDETIFHVL DIISWLD+HSS KNK
Sbjct: 362 HTLLEGEPDETIFHVLGDIISWLDEHSSRKNK 457
>TC205015
Length = 936
Score = 212 bits (540), Expect = 2e-55
Identities = 101/185 (54%), Positives = 133/185 (71%), Gaps = 14/185 (7%)
Frame = +2
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
IEG+S+EL I N+D A RR R AF D+Q +DHCLF+ G++ EE YE NSRG
Sbjct: 380 IEGLSDELNAIARCNLDFAYTRRRVRAAFADMQQQLDHCLFKNAPAGIRTEEWYERNSRG 559
Query: 68 LKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
L++F KSW+P+ P+K V +CHGY DTCTF+FEG+AR +A+SG+ VFA+DYPGFGLS+
Sbjct: 560 LEIFCKSWMPKPGIPIKASVCFCHGYGDTCTFFFEGIARIIAASGYSVFAMDYPGFGLSE 739
Query: 127 GLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGA 173
GLHGYIP F+ LV+DVIEH++KIK G+SMGGA++L +H ++P WDG
Sbjct: 740 GLHGYIPKFDYLVDDVIEHYTKIKARPDLSGLPRFILGQSMGGAVSLKVH*REPNNWDGM 919
Query: 174 ALIAP 178
L+AP
Sbjct: 920 ILVAP 934
>BE059313
Length = 406
Score = 189 bits (479), Expect = 2e-48
Identities = 91/114 (79%), Positives = 97/114 (84%), Gaps = 13/114 (11%)
Frame = +3
Query: 67 GLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
GLK+FSKSW+PE S +K I+ YCHGYADTCTFYFEGVARKLASSG+GVFALDYPGFGLSD
Sbjct: 60 GLKIFSKSWLPESSHLKAIICYCHGYADTCTFYFEGVARKLASSGYGVFALDYPGFGLSD 239
Query: 127 GLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQP 167
GLHGYIPSFE+LVNDVIEHFSKIK GESMGGAIALNIHFKQP
Sbjct: 240 GLHGYIPSFESLVNDVIEHFSKIKEQKKYQDVPSFLLGESMGGAIALNIHFKQP 401
Score = 30.8 bits (68), Expect = 0.90
Identities = 13/14 (92%), Positives = 14/14 (99%)
Frame = -3
Query: 61 YEVNSRGLKVFSKS 74
YEVNSRGLK+FSKS
Sbjct: 59 YEVNSRGLKIFSKS 18
>TC205012 similar to UP|Q8CD59 (Q8CD59) Mus musculus 13 days embryo male
testis cDNA, RIKEN full-length enriched library,
clone:6030422B13 product:SH3 domain protein 1B, full
insert sequence, partial (6%)
Length = 622
Score = 182 bits (463), Expect = 1e-46
Identities = 81/144 (56%), Positives = 112/144 (77%), Gaps = 1/144 (0%)
Frame = +1
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
IEG+SEEL + N+D AP+RR R AF ++ +DH LF+ G++ EE YE NSRG
Sbjct: 178 IEGVSEELNAMASQNLDFAPSRRRVRAAFTEVHQQLDHFLFKTAPPGIRTEEGYERNSRG 357
Query: 68 LKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
L++F KSW+PE P+K + +CHGY TCTF+FEG+A+++ +SG+GV+A+DYPGFGLS+
Sbjct: 358 LEIFCKSWMPESGVPLKAALCFCHGYGSTCTFFFEGIAKRIDASGYGVYAMDYPGFGLSE 537
Query: 127 GLHGYIPSFENLVNDVIEHFSKIK 150
GLHGYIP F++LV+DVIEH++KIK
Sbjct: 538 GLHGYIPKFDDLVDDVIEHYTKIK 609
>TC217654 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, partial (28%)
Length = 792
Score = 174 bits (441), Expect = 5e-44
Identities = 93/177 (52%), Positives = 116/177 (64%), Gaps = 26/177 (14%)
Frame = +1
Query: 185 DMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGT 244
DM+P + ILIG+A VLPK KLVP K+ + E +RD +KRE YNV+ YKDKPRL +
Sbjct: 1 DMVPPKFLTHILIGLANVLPKHKLVPNKD-LAEAAFRDLKKREQTAYNVVAYKDKPRLKS 177
Query: 245 ALELLKATQELEQRLEEVSLPLLVMHGEADI--------------------------ITD 278
A+E+LK T+E+EQRL+EVSLP+ ++HGEA+ +TD
Sbjct: 178 AVEMLKTTEEIEQRLKEVSLPIFILHGEANTR*LNHQ*AKPGITTQAIQTTSGSCTRMTD 357
Query: 279 PSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
PS SKALY+ A DKKL LYKDA+H LLEGEPDE I V DIISWLD+HS T N+
Sbjct: 358 PSVSKALYENASCSDKKLQLYKDAYHALLEGEPDEIITQVFGDIISWLDEHSLTHNQ 528
>TC205013 weakly similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, partial
(47%)
Length = 719
Score = 164 bits (415), Expect = 5e-41
Identities = 92/216 (42%), Positives = 119/216 (54%), Gaps = 13/216 (6%)
Frame = +1
Query: 131 YIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAALIA 177
YIP+F++LV+DVIEHF+KIK G+SMGGAIAL +H K+ WDG L+A
Sbjct: 4 YIPNFDDLVDDVIEHFTKIKARPEVRGLPRFILGQSMGGAIALKVHLKEQNTWDGVILVA 183
Query: 178 PLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYK 237
P+CK A YNV+ Y
Sbjct: 184 PMCK-----------------------------------------------AGYNVISYD 222
Query: 238 DKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLC 297
RL T +ELL ATQE+E +L +VS PLL++HG AD +TDP S+ LY+KA KDK L
Sbjct: 223 HPTRLKTGMELLSATQEIESQLHKVSAPLLILHGAADQVTDPLVSQFLYEKASSKDKTLK 402
Query: 298 LYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTK 333
+Y+ ++H +LEGEPD+ IF V +DIISWLD S K
Sbjct: 403 IYEGSYHGILEGEPDDRIFAVHNDIISWLDFRCSLK 510
>TC220810 similar to PIR|T00552|T00552 lysophospholipase homolog F12L6.8 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 833
Score = 160 bits (406), Expect = 6e-40
Identities = 83/184 (45%), Positives = 116/184 (62%)
Frame = +2
Query: 151 GESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVP 210
GESMGGA+AL +H K+P WDGA L+AP+CK AE+M P+ +V +L +++V P ++VP
Sbjct: 5 GESMGGAVALLLHRKKPEYWDGAILVAPMCKIAEEMKPNTMVISVLSALSRVFPSWRIVP 184
Query: 211 QKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMH 270
+ + + ++ + RE N YK PRL TA ELL+ + E+EQ L EVSLP +V+H
Sbjct: 185 TPDII-DLAFKVPKVREEIRANRYCYKGNPRLRTAYELLRVSTEIEQSLHEVSLPFIVLH 361
Query: 271 GEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
GE D +TD + SK LY +A DK L Y + +H LL GEP + + V DII W+D S
Sbjct: 362 GEEDQVTDKAVSKQLYDEAASSDKTLKSYPEMWHGLLYGEPPQNLQIVFSDIIGWIDQKS 541
Query: 331 STKN 334
N
Sbjct: 542 RYGN 553
>TC217616 similar to GB|AAM63479.1|21554372|AY086477 phospholipase-like
protein {Arabidopsis thaliana;} , partial (95%)
Length = 1400
Score = 159 bits (401), Expect = 2e-39
Identities = 96/280 (34%), Positives = 145/280 (51%), Gaps = 14/280 (5%)
Frame = +3
Query: 61 YEVNSRGLKVFSKSWIP-EKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDY 119
+ N RGLK+F++ W P + G + HGY ++ + A A +GF ALD+
Sbjct: 282 FVTNPRGLKLFTQWWTPLPPKTIIGTLAVVHGYTGESSWLLQLTAVHFAKAGFATCALDH 461
Query: 120 PGFGLSDGLHGYIPSFENLVNDVIEHFSKIKG------------ESMGGAIALNIHFKQ- 166
G G SDGL +IP +V+D I F + ES+GGAIAL I ++
Sbjct: 462 QGHGFSDGLVAHIPDINPVVDDCITFFENFRSRFDPSLPSFLYAESLGGAIALLITLRRR 641
Query: 167 PTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKR 226
W G L +C + P W ++ L VA V+P ++VP + + E ++ KR
Sbjct: 642 EMLWSGVILNGAMCGISAKFKPPWPLEHFLSVVAAVIPTWRVVPTRGSIPEVSFKVEWKR 821
Query: 227 ELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALY 286
LA + +PR TA ELL+ +EL+ R EEV +PLLV HG D++ DP+ + L+
Sbjct: 822 RLALASPRRTVARPRAATAQELLRICRELQGRYEEVEVPLLVAHGGDDVVCDPACVEELH 1001
Query: 287 QKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWL 326
+A KDK L +Y +H ++ GEP+E + V D++ WL
Sbjct: 1002ARAASKDKTLKIYPGMWHQMV-GEPEENVELVFGDMLEWL 1118
>TC217653 similar to UP|Q9LF16 (Q9LF16) Lipase-like protein, partial (34%)
Length = 548
Score = 130 bits (328), Expect(2) = 3e-38
Identities = 59/97 (60%), Positives = 73/97 (74%)
Frame = +3
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
I G+ EL+ I +NMDE ARR AREAFK+IQL IDH LF+ P DG+KMEE YE NS+G
Sbjct: 108 IPGVDRELKKILKANMDEVGARRRAREAFKNIQLGIDHILFKTPCDGIKMEESYEKNSKG 287
Query: 68 LKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVA 104
L++F KSW+P S K V+YCHGY DTC+F+FE +A
Sbjct: 288 LEIFCKSWLPSASKPKAAVFYCHGYGDTCSFFFERIA 398
Score = 45.8 bits (107), Expect(2) = 3e-38
Identities = 24/43 (55%), Positives = 29/43 (66%), Gaps = 13/43 (30%)
Frame = +2
Query: 131 YIPSFENLVNDVIEHFSKIK-------------GESMGGAIAL 160
YIPSF+ LV+DVIEH+SKIK G+SMGGA+AL
Sbjct: 398 YIPSFDGLVDDVIEHYSKIKENPEFHSLPSFLFGQSMGGAVAL 526
>TC228275 weakly similar to UP|Q9M3D0 (Q9M3D0) Lipase-like protein, partial
(25%)
Length = 1153
Score = 153 bits (387), Expect = 9e-38
Identities = 94/283 (33%), Positives = 143/283 (50%), Gaps = 23/283 (8%)
Frame = +2
Query: 69 KVFSKSWIP---EKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLS 125
K+F++S++P + +K V+ HGY + F+ + A+ G+ VFA D G G S
Sbjct: 110 KIFTQSFLPLNLQPHQVKATVFMTHGYGSDTGWLFQKICINFATWGYAVFAADLLGHGRS 289
Query: 126 DGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFK-QPTAWD 171
DGL Y+ + + + F ++ GESMGG L ++FK +P W
Sbjct: 290 DGLQCYLGDMDKIAATSLSFFLHVRNSHPYKNLPAFLFGESMGGLATLLMYFKSEPDTWT 469
Query: 172 GAALIAPLCKFAEDMIP---HWLVKQILIGVAKV---LPKTKLVPQKEEVKENIYRDARK 225
G APL EDM P H + +L G+A +P K+V + RD K
Sbjct: 470 GLMFSAPLFVIPEDMKPSRVHLFMYGLLFGLADTWAAMPDNKMVGKA-------IRDPEK 628
Query: 226 RELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKAL 285
++ N Y PR+GT ELL+ TQ ++ +V+ P HG +D +T PS+SK L
Sbjct: 629 LKVIASNPRRYTGPPRVGTMRELLRVTQYVQDNFSKVTTPFFTAHGTSDGVTCPSSSKLL 808
Query: 286 YQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDD 328
Y+K +DK L LY +H+L++GEPDE+ VL D+ W+D+
Sbjct: 809 YEKGSSEDKTLKLYDGMYHSLIQGEPDESANLVLGDMREWIDE 937
>BI968079 weakly similar to PIR|H86244|H86 lysophospholipase homolog
25331-24357 [imported] - Arabidopsis thaliana, partial
(49%)
Length = 754
Score = 144 bits (363), Expect = 6e-35
Identities = 76/188 (40%), Positives = 119/188 (62%), Gaps = 8/188 (4%)
Frame = -1
Query: 147 SKIKGESMGGAIALNIHF--------KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIG 198
S + GESMG I+L IH QP + GA L+AP+C ++++ P W + QIL
Sbjct: 712 SFLYGESMGAXISLLIHLVNSETEPKSQP--FQGAVLVAPMCXISDNVRPKWPIPQILTF 539
Query: 199 VAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQR 258
+++ P +VP + + +++ D K+ +A N L Y+ KPRLGT +ELL+ T L +R
Sbjct: 538 LSRFFPTLPIVPTPDLLYKSVKVD-HKKVIADMNPLRYRGKPRLGTVVELLRVTDLLSRR 362
Query: 259 LEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHV 318
L +VSLP +V+HG AD++TDP+ S+ LY++A+ DK + +Y++ H+LL GE DE + V
Sbjct: 361 LCDVSLPFIVLHGSADVVTDPNVSRELYREARSDDKTIKVYEEMMHSLLFGETDENVEIV 182
Query: 319 LDDIISWL 326
+DI+ WL
Sbjct: 181 RNDILEWL 158
>BM188435
Length = 430
Score = 140 bits (352), Expect = 1e-33
Identities = 68/142 (47%), Positives = 96/142 (66%)
Frame = +3
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K P+ WDGA L+AP+CK +E + PH +V IL V ++PK K+VP K+ + ++ ++D
Sbjct: 6 KDPSFWDGAVLVAPMCKISEKVKPHPVVVNILTKVEDIIPKWKIVPTKDVI-DSAFKDPA 182
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KRE N L Y+DKPRL TALE+L+ + LE L +V+LP V+HGEAD +TDP S+A
Sbjct: 183 KRERIRKNKLIYQDKPRLKTALEMLRISMSLEDSLYKVTLPFFVLHGEADTVTDPEVSRA 362
Query: 285 LYQKAKVKDKKLCLYKDAFHTL 306
LY++A KDK + LY +H L
Sbjct: 363 LYERASSKDKTIKLYPGMWHGL 428
>BU548111
Length = 689
Score = 106 bits (265), Expect(2) = 3e-33
Identities = 50/92 (54%), Positives = 69/92 (74%)
Frame = -1
Query: 236 YKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKK 295
+++K L T +ELL ATQE+E L +VS PLL++HG D++TDP S+ LY+KA KDK
Sbjct: 521 WQEKSGLKTGMELLSATQEIESLLHKVSAPLLILHGADDLVTDPLVSQFLYEKASSKDKT 342
Query: 296 LCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
L +Y+ ++H +LEGEPD+ IF V +DIISWLD
Sbjct: 341 LEIYEGSYHGILEGEPDDRIFAVHNDIISWLD 246
Score = 53.1 bits (126), Expect(2) = 3e-33
Identities = 25/62 (40%), Positives = 38/62 (60%)
Frame = -2
Query: 167 PTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKR 226
P WDG L+AP+CK AE M P V +L ++KV+PK KL P + ++ +R+ KR
Sbjct: 688 PNTWDGVILVAPMCKVAEGMXPPMAVXXVLNLLSKVMPKAKLFPHR-DLSALTFREPGKR 512
Query: 227 EL 228
++
Sbjct: 511 KV 506
>TC206888 similar to UP|Q8LEE3 (Q8LEE3) Lysophospholipase-like protein,
partial (83%)
Length = 1499
Score = 118 bits (296), Expect = 3e-27
Identities = 95/287 (33%), Positives = 135/287 (46%), Gaps = 25/287 (8%)
Frame = +1
Query: 66 RGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLS 125
R +F +SW P +KGI+ HG + Y + AR+L S FGV+A+D+ G G S
Sbjct: 397 RNNALFCRSWFPVAGDVKGILIIIHGLNEHSGRYAD-FARQLTSCNFGVYAMDWIGHGGS 573
Query: 126 DGLHGYIPSFENLVNDVIEHFSKIK-----------GESMGGAIALNI--HFKQPTAWDG 172
DGLHGY+PS +++V D KI+ G S GGA+ L H +G
Sbjct: 574 DGLHGYVPSLDHVVADTGAFLEKIRSENPGIPCFLFGHSTGGAVVLKAASHPHIEVMVEG 753
Query: 173 AALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRE----- 227
L +P + VK V V P LV + + ++ A KR
Sbjct: 754 IILTSPALR----------VKPAHPIVGAVAPIFSLVAPRFQ-----FKGANKRGIPVSR 888
Query: 228 -----LAPY-NVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSA 281
LA Y + L Y R+ T E+L+ + L + V++P V+HG AD +TDP A
Sbjct: 889 DPAALLAKYSDPLVYTGPIRVRTGHEILRISSYLMRNFNSVTVPFFVLHGTADKVTDPLA 1068
Query: 282 SKALYQKAKVKDKKLCLYKDAFHTLL-EGEPDETIFHVLDDIISWLD 327
S+ LY KA K K + LY H LL E E +E + DII+W++
Sbjct: 1069SQDLYDKAASKFKDIKLYDGFLHDLLFEPEREE----IAQDIINWME 1197
>BI321608
Length = 363
Score = 82.4 bits (202), Expect(2) = 4e-26
Identities = 33/52 (63%), Positives = 49/52 (93%)
Frame = +2
Query: 102 GVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIKGES 153
G+A+++ +SG+GV+A+DYP FGLS+GLHGYIP F++LV+DVIEH++KIKG++
Sbjct: 152 GIAKRIDASGYGVYAMDYPFFGLSEGLHGYIPKFDDLVDDVIEHYTKIKGKT 307
Score = 53.5 bits (127), Expect(2) = 4e-26
Identities = 22/43 (51%), Positives = 31/43 (71%), Gaps = 1/43 (2%)
Frame = +1
Query: 54 GVKMEEIYEVNSRGLKVFSKSWIPEKS-PMKGIVYYCHGYADT 95
G++ EE YE NSRGL++F K+W+PE P+K + +CHGY T
Sbjct: 37 GIRTEEWYERNSRGLEIFCKNWMPEPGVPLKAALCFCHGYCQT 165
>TC230682
Length = 798
Score = 101 bits (251), Expect = 5e-22
Identities = 47/87 (54%), Positives = 63/87 (72%)
Frame = +1
Query: 247 ELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFHTL 306
ELL+ T+E+E ++ +VS PLL++HG D +TDP SK LY++A KDK L LY+ +H +
Sbjct: 1 ELLRTTKEIESQVHKVSAPLLILHGAEDKVTDPLVSKFLYERASSKDKTLKLYEGGYHCI 180
Query: 307 LEGEPDETIFHVLDDIISWLDDHSSTK 333
LEGEPD+ IF V DDI+SWLD S K
Sbjct: 181 LEGEPDDRIFAVHDDIVSWLDFRCSIK 261
>CB063434
Length = 408
Score = 95.5 bits (236), Expect = 3e-20
Identities = 47/121 (38%), Positives = 69/121 (56%), Gaps = 13/121 (10%)
Frame = +1
Query: 61 YEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYP 120
Y NSRG+++F+ W+P SP KG+V+ CHGY C+ + +LA + + VF +DY
Sbjct: 46 YRRNSRGVQLFTCKWLPFSSP-KGLVFLCHGYGMECSGFMRECGVRLACAKYAVFGMDYE 222
Query: 121 GFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQP 167
G G S+G YI F+N+VND + F + GESMGGA++L +H K P
Sbjct: 223 GHGRSEGARCYIKKFDNIVNDCYDFFKSVSELQEYKAKARFLYGESMGGAVSLLLHKKDP 402
Query: 168 T 168
+
Sbjct: 403 S 405
>TC230840 weakly similar to PIR|T00552|T00552 lysophospholipase homolog
F12L6.8 - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (33%)
Length = 460
Score = 89.7 bits (221), Expect = 2e-18
Identities = 36/95 (37%), Positives = 61/95 (63%)
Frame = +3
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGV 114
+K +E Y NSRGLK+F+ W+P K +++ CHGYA C+ + +LA +GF V
Sbjct: 138 IKYDEEYVQNSRGLKLFACRWLPANGSPKALIFLCHGYAMECSITMKSTGTRLAKAGFAV 317
Query: 115 FALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI 149
+ +DY G G S+G+ G + +F+ +++D +HF++I
Sbjct: 318 YGIDYEGHGKSEGVPGLVMNFDFVIDDCSQHFTRI 422
>TC222461 similar to UP|Q8RZT4 (Q8RZT4) Phospholipase-like protein, partial
(9%)
Length = 627
Score = 82.4 bits (202), Expect = 3e-16
Identities = 43/108 (39%), Positives = 65/108 (59%), Gaps = 13/108 (12%)
Frame = +2
Query: 134 SFENLVNDVIEHFSKIKG-------------ESMGGAIALNIHFKQPTAWDGAALIAPLC 180
+F++LV+DVIEHF+K K +SMGGAIAL +H K+ WDG L+AP+C
Sbjct: 2 NFDDLVDDVIEHFTKFKARPEVRGLPRFILRQSMGGAIALKVHLKEQNTWDGVILVAPMC 181
Query: 181 KFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKREL 228
K A M+P + ++L ++ V+PK KL P K ++ +R KR++
Sbjct: 182 KIA*GMLPPTALLRVLNLLSNVMPKAKLFPHK-DLSALTFRQPGKRKV 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,397,225
Number of Sequences: 63676
Number of extensions: 191773
Number of successful extensions: 873
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 840
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 846
length of query: 336
length of database: 12,639,632
effective HSP length: 98
effective length of query: 238
effective length of database: 6,399,384
effective search space: 1523053392
effective search space used: 1523053392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146563.12


