

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
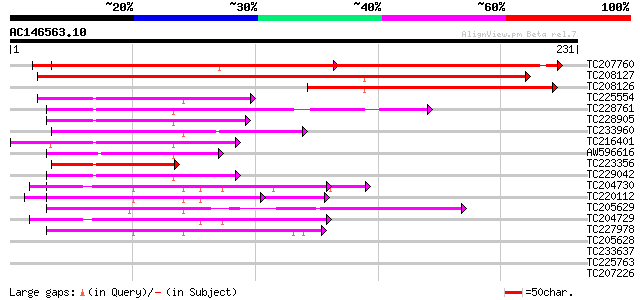
Query= AC146563.10 + phase: 0
(231 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207760 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 397 e-111
TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 334 2e-92
TC208126 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8,... 130 5e-31
TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 50 9e-07
TC228761 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {A... 50 1e-06
TC228905 weakly similar to UP|Q7XIV3 (Q7XIV3) Zinc finger and C2... 49 3e-06
TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%) 48 3e-06
TC216401 weakly similar to UP|Q9LP65 (Q9LP65) T1N15.21, partial ... 47 1e-05
AW596616 45 2e-05
TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {A... 45 4e-05
TC229042 similar to UP|Q9LVH4 (Q9LVH4) GTPase activating protein... 44 6e-05
TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, par... 43 1e-04
TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial ... 43 1e-04
TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050),... 42 2e-04
TC204729 weakly similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 prote... 42 2e-04
TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmi... 41 5e-04
TC205628 similar to UP|O48645 (O48645) CLB1, partial (38%) 39 0.003
TC233637 38 0.004
TC225763 38 0.004
TC207226 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {A... 37 0.010
>TC207760 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial (40%)
Length = 1835
Score = 397 bits (1021), Expect = e-111
Identities = 200/216 (92%), Positives = 205/216 (94%)
Frame = +3
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G G GNGWIELVLIEGRDLVAAD+RGTSDP+VRVHYGNFKK+TKVIYKTL PQWNQTLEF
Sbjct: 894 GLGLGNGWIELVLIEGRDLVAADVRGTSDPFVRVHYGNFKKKTKVIYKTLNPQWNQTLEF 1073
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRK 129
PDDGS LMLYVKDHNALLPTSSIGECVVEYQRLPPNQ ADKWIPLQGVKRGEIHIQITRK
Sbjct: 1074 PDDGSQLMLYVKDHNALLPTSSIGECVVEYQRLPPNQTADKWIPLQGVKRGEIHIQITRK 1253
Query: 130 VPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQE 189
VPEMQKRQS+DSEPSLSKLHQIP QIKQMMIKFRS IEDGNLEGLS TLSELETLEDTQE
Sbjct: 1254 VPEMQKRQSLDSEPSLSKLHQIPNQIKQMMIKFRSFIEDGNLEGLSATLSELETLEDTQE 1433
Query: 190 GYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRR 225
GY+ QLETEQMLLLSKIKELGQEIINS SPSLSRR
Sbjct: 1434 GYIVQLETEQMLLLSKIKELGQEIINS--SPSLSRR 1535
Score = 75.5 bits (184), Expect = 2e-14
Identities = 38/118 (32%), Positives = 65/118 (54%), Gaps = 1/118 (0%)
Frame = +3
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
I + ++EG+DL A D G DPY+++ YG ++T+ ++ T P WNQ EF + G
Sbjct: 528 INVTVVEGKDLAAKDKSGKFDPYIKLQYGKVVQKTRTVH-TPNPVWNQMFEFDEIGGGEY 704
Query: 78 LYVKDHN-ALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQ 134
L +K + + +IG V + L + D WIPL+ V+ GE+ +QI+ + + +
Sbjct: 705 LKLKGFSEEIFGDENIGSAHVNLEGLVEGSVRDVWIPLERVRSGELRLQISIRADDQE 878
>TC208127 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial
(23%)
Length = 845
Score = 334 bits (857), Expect = 2e-92
Identities = 167/202 (82%), Positives = 185/202 (90%), Gaps = 1/202 (0%)
Frame = +3
Query: 12 GSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPD 71
GS NGWIELV+IE RDL+AADLRGTSDPYVRV+YGN KKRTKVI+KTL P+WNQTLEF D
Sbjct: 240 GSTNGWIELVVIEARDLIAADLRGTSDPYVRVNYGNSKKRTKVIHKTLNPRWNQTLEFLD 419
Query: 72 DGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVP 131
DGSPL+L+VKDHNALLP SSIGE VVEYQRLPPNQM+DKWIPLQGVK GEIHIQITRKVP
Sbjct: 420 DGSPLILHVKDHNALLPESSIGEGVVEYQRLPPNQMSDKWIPLQGVKSGEIHIQITRKVP 599
Query: 132 EMQKRQSMDSEP-SLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQEG 190
EMQ R ++DS+P SLSK HQIPTQ+++MM KFRS IED NLEGL+TTLSELE+LEDTQEG
Sbjct: 600 EMQTRHTLDSQPSSLSKSHQIPTQMREMMKKFRSLIEDENLEGLTTTLSELESLEDTQEG 779
Query: 191 YVAQLETEQMLLLSKIKELGQE 212
Y+ QLETEQMLLLSKI ELG+E
Sbjct: 780 YITQLETEQMLLLSKINELGRE 845
>TC208126 weakly similar to UP|Q93ZM0 (Q93ZM0) AT3g18370/MYF24_8, partial
(8%)
Length = 574
Score = 130 bits (327), Expect = 5e-31
Identities = 71/103 (68%), Positives = 81/103 (77%), Gaps = 1/103 (0%)
Frame = +3
Query: 122 IHIQITRKVPEMQKRQSMDSEP-SLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSE 180
IHI +RKVP Q R + +P SLSK HQIPTQ+++MM KFRS IED NLEGL+TTLSE
Sbjct: 3 IHIXXSRKVPXXQSRHTXGCQPCSLSKSHQIPTQMREMMKKFRSLIEDENLEGLTTTLSE 182
Query: 181 LETLEDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLS 223
LE+LEDTQEGY+ QLETEQMLL SK ELG+EIINSS S S
Sbjct: 183 LESLEDTQEGYITQLETEQMLLPSKRNELGREIINSSSRASSS 311
>TC225554 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (95%)
Length = 1383
Score = 50.1 bits (118), Expect = 9e-07
Identities = 31/91 (34%), Positives = 47/91 (51%), Gaps = 2/91 (2%)
Frame = +3
Query: 12 GSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF-- 69
G G +++++++G+ LV D + TSDPYV + GN +TKVI L P WN+ L F
Sbjct: 288 GEQLGLLKVIVVQGKRLVIRDFK-TSDPYVVLKLGNQTAKTKVINSCLNPVWNEELNFTL 464
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQ 100
+ L L V D + L +G + Q
Sbjct: 465 TEPLGVLNLEVFDKDLLKADDKMGNAFLNLQ 557
>TC228761 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {Arabidopsis
thaliana;} , partial (48%)
Length = 729
Score = 49.7 bits (117), Expect = 1e-06
Identities = 41/159 (25%), Positives = 73/159 (45%), Gaps = 2/159 (1%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G +++ +I+G +L D++ +SDPYV + G +T +I L P WN+ L P+
Sbjct: 34 GMLKVKVIKGTNLAIRDIK-SSDPYVVLSLGQQTVQTTIIRSNLNPVWNEEYMLSVPEHY 210
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEM 133
+ L V DH+ +GE ++ Q L + MA + G ++QI + +
Sbjct: 211 GQMKLKVFDHDTFSADDIMGEADIDLQSLITSAMAFGDAGMFG------NMQIGKWLKSD 372
Query: 134 QKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLE 172
DS ++ + ++KQMM +E G L+
Sbjct: 373 DNALIEDSTVNI-----VDGKVKQMMSLKLQDVESGELD 474
>TC228905 weakly similar to UP|Q7XIV3 (Q7XIV3) Zinc finger and C2 domain
protein-like, partial (85%)
Length = 819
Score = 48.5 bits (114), Expect = 3e-06
Identities = 29/85 (34%), Positives = 45/85 (52%), Gaps = 2/85 (2%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G ++L + G +L D R TSDPYV V+ G+ K +T+V+ P WN+ TL D
Sbjct: 57 GLLKLRIKRGVNLAIRDAR-TSDPYVVVNMGDQKLKTRVVKNNCNPDWNEELTLSVKDVK 233
Query: 74 SPLMLYVKDHNALLPTSSIGECVVE 98
+P+ L V D + +GE ++
Sbjct: 234 TPIHLTVYDKDTFSVDDKMGEAEID 308
>TC233960 similar to UP|OPSD_SEPOF (O16005) Rhodopsin, partial (10%)
Length = 862
Score = 48.1 bits (113), Expect = 3e-06
Identities = 34/109 (31%), Positives = 51/109 (46%), Gaps = 5/109 (4%)
Frame = +2
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF-----PDD 72
+++ +I L AAD GTSDP+V G+ +T V K L P WN+ L+F
Sbjct: 89 LKIRVISASSLQAADRGGTSDPFVHFKQGHMGVKTTVKNKELNPVWNEDLQFGIYEKDAQ 268
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
L + V D + +L IG V +P NQ + + L G + G+
Sbjct: 269 TGTLSIEVLDKD-MLSDDCIGTYKVSLSEVPHNQPREYDVHLTGGQSGK 412
>TC216401 weakly similar to UP|Q9LP65 (Q9LP65) T1N15.21, partial (82%)
Length = 944
Score = 46.6 bits (109), Expect = 1e-05
Identities = 30/98 (30%), Positives = 51/98 (51%), Gaps = 4/98 (4%)
Frame = +3
Query: 1 MIHFDNNDQGSGSGN--GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKT 58
++HF + S N G + + + +G +L D+ +SDPYV + G K +T+V+ K
Sbjct: 51 LLHFIPHIYFSSMENLLGLLRIHVEKGVNLAIRDVV-SSDPYVVIKMGKQKLKTRVVNKN 227
Query: 59 LTPQWNQ--TLEFPDDGSPLMLYVKDHNALLPTSSIGE 94
L P+WN TL D +P+ L+V D + +G+
Sbjct: 228 LNPEWNDDLTLSISDPHAPIHLHVYDKDTFSMDDKMGD 341
>AW596616
Length = 337
Score = 45.4 bits (106), Expect = 2e-05
Identities = 25/74 (33%), Positives = 41/74 (54%), Gaps = 2/74 (2%)
Frame = +2
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G + + + +G +L D+ T DPYV + G K +T+V+ K L P+WN TL D
Sbjct: 14 GLLRIHVEKGVNLAIRDVVST-DPYVVIKMGKSKLKTRVVDKNLNPEWNDDLTLSISDPH 190
Query: 74 SPLMLYVKDHNALL 87
+P+ L+V D + +
Sbjct: 191APIHLHVYDKDTFI 232
>TC223356 similar to GB|AAO44032.1|28466847|BT004766 At5g47710 {Arabidopsis
thaliana;} , partial (96%)
Length = 705
Score = 44.7 bits (104), Expect = 4e-05
Identities = 21/52 (40%), Positives = 34/52 (65%)
Frame = +3
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
+++++++G+ LV D + +SDPYV V GN +T+VI L P WN+ L F
Sbjct: 87 LKVIVVQGKRLVIRDFK-SSDPYVVVKLGNQTAKTRVIRCCLNPVWNEELNF 239
>TC229042 similar to UP|Q9LVH4 (Q9LVH4) GTPase activating protein-like,
partial (95%)
Length = 783
Score = 43.9 bits (102), Expect = 6e-05
Identities = 26/81 (32%), Positives = 43/81 (52%), Gaps = 2/81 (2%)
Frame = +3
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPDDG 73
G + + + G +L D+R +SDPYV + N K +T+VI K + P+WN+ TL +
Sbjct: 33 GLLRVRVKRGVNLAVRDVR-SSDPYVVIKMYNQKLKTRVIKKDVNPEWNEDLTLSVINPN 209
Query: 74 SPLMLYVKDHNALLPTSSIGE 94
+ L V DH+ +G+
Sbjct: 210 HKIKLTVYDHDTFSKDDKMGD 272
>TC204730 similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, partial (97%)
Length = 2084
Score = 43.1 bits (100), Expect = 1e-04
Identities = 36/149 (24%), Positives = 62/149 (41%), Gaps = 17/149 (11%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFK---KRTKVIYKTLTPQWNQTLEF--- 69
G + + ++ L DL G SDPYV++ K K+T V YK L P+WN+
Sbjct: 979 GILHVKVVRAEKLKKKDLLGASDPYVKLKLTEEKLPSKKTTVKYKNLNPEWNEEFNIVVK 1158
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQ----------MADKWIPLQGVKR 119
+ L L V D + +G V+ + + P++ D P R
Sbjct: 1159 DPESQVLELTVYDWEQIGKHDKMGMNVIPLKEITPDEPKAVTLNLLKTMDPNDPENAKSR 1338
Query: 120 GEIHIQITRK-VPEMQKRQSMDSEPSLSK 147
G++ +++ K E + QS + ++ K
Sbjct: 1339 GQLTVEVLYKPFKEDELPQSAEDSNAIEK 1425
Score = 42.0 bits (97), Expect = 2e-04
Identities = 29/131 (22%), Positives = 64/131 (48%), Gaps = 8/131 (6%)
Frame = +1
Query: 9 QGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLE 68
+G+ + G + +++ E D+ + ++PYVR+ + +++TK + K P+W ++ +
Sbjct: 1432 EGTPASGGLLVIIVHEAEDVEG---KHHTNPYVRLLFKGEERKTKHVKKNRDPRWGESFQ 1602
Query: 69 FPDDGSPL--MLYVKDHNA------LLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRG 120
F + P LYV+ +A L P S+G ++ + N+ ++ L + G
Sbjct: 1603 FMLEEPPTNERLYVEVQSASSKLGLLHPKESLGYVDIKLSDVVTNKRINEKYHLIDSRNG 1782
Query: 121 EIHIQITRKVP 131
I I++ + P
Sbjct: 1783 RIQIELQWRTP 1815
>TC220112 similar to UP|Q7XA06 (Q7XA06) Synaptotagmin C, partial (76%)
Length = 1867
Score = 43.1 bits (100), Expect = 1e-04
Identities = 32/131 (24%), Positives = 61/131 (46%), Gaps = 7/131 (5%)
Frame = +2
Query: 7 NDQGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQT 66
+D G G + +V+ E + V D ++P+ + + KKRTK + KT P+WN+
Sbjct: 1346 SDDEVQEGAGLLSIVIQEAEE-VEGDHH--NNPFAVLTFRGEKKRTKTMKKTRHPRWNEE 1516
Query: 67 LEFPDDGSPL-------MLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKR 119
+F + PL ++ + + + LP S+G + + + N + L +
Sbjct: 1517 FQFMLEEPPLHEKIHIEVMSKRKNFSFLPKESLGHVEINLRDVVHNGRINDKYHLINSRN 1696
Query: 120 GEIHIQITRKV 130
G +H++I KV
Sbjct: 1697 GVMHVEIRWKV 1729
Score = 40.4 bits (93), Expect = 7e-04
Identities = 28/95 (29%), Positives = 45/95 (46%), Gaps = 6/95 (6%)
Frame = +2
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFK---KRTKVIYKTLTPQWNQTLEF--- 69
G + + ++ + L+ DL GTSDPYV++ K K+T V K L P+WN+ +
Sbjct: 878 GILHVNVVRAQKLLKMDLLGTSDPYVKLSLTGDKLPAKKTTVKRKNLNPEWNEKFKIVVK 1057
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPP 104
L L V D + + +G +V + L P
Sbjct: 1058DPQSQVLQLQVYDWDKVGGHDKLGMQLVPLKVLKP 1162
>TC205629 similar to UP|Q9LEX1 (Q9LEX1) CaLB protein (At3g61050), partial
(92%)
Length = 1766
Score = 42.4 bits (98), Expect = 2e-04
Identities = 46/175 (26%), Positives = 74/175 (42%), Gaps = 4/175 (2%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-FKKRTKVIYKTLTPQWNQTLEF---PD 71
G + L +++ L ++ G SDPYV VH FK +TKVI L P WN+ E
Sbjct: 697 GKLALTVVKATALKNMEMIGKSDPYVVVHIRPLFKYKTKVIDNNLNPTWNEKFELIAEDK 876
Query: 72 DGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVP 131
+ L+L V D IG+ DK + + + ++ IQ T K
Sbjct: 877 ETQSLILEVLD-------KDIGQ--------------DKRLGIAQLPLIDLEIQ-TEKEI 990
Query: 132 EMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLED 186
E++ S+D+ K + +K M +F + + LE L E + L++
Sbjct: 991 ELRLLPSLDTLKVKDKKDRGTLTVKVMYYQFNKEEQLVALEAEKKILEERKKLKE 1155
>TC204729 weakly similar to UP|Q8LQF0 (Q8LQF0) OJ1529_G03.6 protein, partial
(35%)
Length = 944
Score = 42.0 bits (97), Expect = 2e-04
Identities = 29/131 (22%), Positives = 64/131 (48%), Gaps = 8/131 (6%)
Frame = +3
Query: 9 QGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLE 68
+G+ + G + +++ E D+ + ++PYVR+ + +++TK + K P+W ++ +
Sbjct: 234 EGTPASGGLLVIIVHEAEDVEG---KHHTNPYVRLLFKGEERKTKHVKKNRDPRWGESFQ 404
Query: 69 FPDDGSPL--MLYVKDHNA------LLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRG 120
F + P LYV+ +A L P S+G ++ + N+ ++ L + G
Sbjct: 405 FMLEEPPTNERLYVEVQSASSKLGLLHPKESLGYVDIKLSDVVTNKRINEKYHLIDSRNG 584
Query: 121 EIHIQITRKVP 131
I I++ + P
Sbjct: 585 RIQIELQWRTP 617
>TC227978 similar to GB|CAE85115.1|39918793|AJ617630 synaptotagmin
{Arabidopsis thaliana;} , partial (72%)
Length = 1470
Score = 40.8 bits (94), Expect = 5e-04
Identities = 34/130 (26%), Positives = 57/130 (43%), Gaps = 16/130 (12%)
Frame = +2
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFK---KRTKVIYKTLTPQWNQTLEF--- 69
G + + +++ L DL G SDPYV++ K K+T V +K L PQWN+
Sbjct: 377 GILHVKVLQAMKLKKKDLLGASDPYVKLKLTEDKWPSKKTTVKHKNLNPQWNEEFNIVVK 556
Query: 70 PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL---------QGVK-R 119
D + + V D + +G V+ + + P + + L Q K R
Sbjct: 557 DPDSQVIEINVYDWEQVGKHDKMGMNVIPLKEVSPEETKRFTLDLLKNMDPNDAQNEKSR 736
Query: 120 GEIHIQITRK 129
G+I +++T K
Sbjct: 737 GQIVVELTYK 766
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/131 (21%), Positives = 59/131 (44%), Gaps = 7/131 (5%)
Frame = +2
Query: 9 QGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLE 68
+G+ +G G + +++ E +D+ + ++P+VR+ + +K+TK I K P+W +
Sbjct: 830 EGTPAGGGLLVVIVHEAQDVEG---KYHTNPHVRLIFRGDEKKTKRIKKNRDPRWEDEFQ 1000
Query: 69 FPDDGSPL-------MLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
F D P ++ N L S+G + + N+ ++ L K G
Sbjct: 1001FMVDEPPTNDRLHVEVVSTSSRNLLHQKESLGYIDINLGDVVANKRINEKYHLIDSKNGR 1180
Query: 122 IHIQITRKVPE 132
+ I++ + E
Sbjct: 1181LQIELQWRTSE 1213
>TC205628 similar to UP|O48645 (O48645) CLB1, partial (38%)
Length = 814
Score = 38.5 bits (88), Expect = 0.003
Identities = 21/51 (41%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Frame = +2
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-FKKRTKVIYKTLTPQWNQ 65
G + L +++ L ++ G SDPYV VH FK +TKVI L P WN+
Sbjct: 98 GKLALTVVKATALKNMEMIGKSDPYVVVHIRPLFKYKTKVIDNNLNPIWNE 250
>TC233637
Length = 426
Score = 37.7 bits (86), Expect = 0.004
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 4/73 (5%)
Frame = +1
Query: 16 GWIELVLIEGRDLVAADLRG--TSDPYVRVHYGNFKKRTKVIYKTLTPQWNQ--TLEFPD 71
G +EL ++ R+L+ R T+D Y YGN RT+ + TL+P+WN+ T E D
Sbjct: 181 GILELGILSARNLLPMKAREGRTTDAYCVAKYGNKWVRTRTLLDTLSPRWNEQYTWEVHD 360
Query: 72 DGSPLMLYVKDHN 84
+ + + V D++
Sbjct: 361 PCTVITVGVFDNH 399
>TC225763
Length = 847
Score = 37.7 bits (86), Expect = 0.004
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 3/92 (3%)
Frame = +2
Query: 9 QGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKT-LTPQWNQTL 67
Q S G +E+VLI + + D + DPYV + Y +K++ V PQWN++
Sbjct: 137 QFSKMPRGTLEVVLISAKGIDDNDFLSSIDPYVILTYRAQEKKSTVQEDAGSKPQWNESF 316
Query: 68 EF--PDDGSPLMLYVKDHNALLPTSSIGECVV 97
F D S L L + D + +GE +
Sbjct: 317 LFTVSDSASELNLKIMDKDNFSQDDCLGEATI 412
>TC207226 similar to GB|AAP68346.1|31711980|BT008907 At1g05500 {Arabidopsis
thaliana;} , partial (16%)
Length = 621
Score = 36.6 bits (83), Expect = 0.010
Identities = 24/81 (29%), Positives = 41/81 (49%), Gaps = 3/81 (3%)
Frame = +1
Query: 49 KKRTKVIYKTLTPQWNQTLEF-PDDGSPLMLYVK--DHNALLPTSSIGECVVEYQRLPPN 105
K +T+V+ +L P WNQT +F +DG ML V+ DH+ +G C++ R+
Sbjct: 10 KNKTRVVNDSLNPVWNQTFDFVVEDGLHDMLIVEVWDHDT-FGKDYMGRCILTLTRVILE 186
Query: 106 QMADKWIPLQGVKRGEIHIQI 126
+ L G K G +++ +
Sbjct: 187 GEYKERFVLDGAKSGFLNLHL 249
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,190,684
Number of Sequences: 63676
Number of extensions: 114832
Number of successful extensions: 537
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 534
length of query: 231
length of database: 12,639,632
effective HSP length: 94
effective length of query: 137
effective length of database: 6,654,088
effective search space: 911610056
effective search space used: 911610056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146563.10


