

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.3 + phase: 0
(262 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
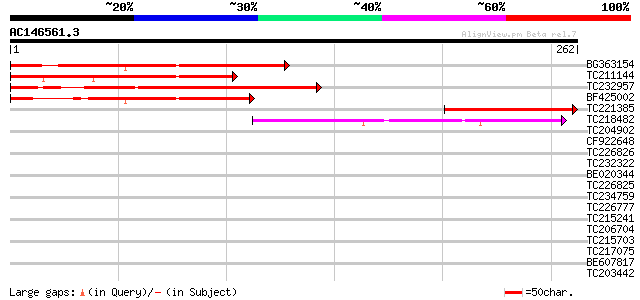
Score E
Sequences producing significant alignments: (bits) Value
BG363154 147 5e-36
TC211144 145 2e-35
TC232957 142 2e-34
BF425002 112 2e-25
TC221385 109 1e-24
TC218482 45 3e-05
TC204902 UP|NO51_SOYBN (P04671) Nodulin C51 precursor, complete 34 0.077
CF922648 32 0.38
TC226826 similar to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydra... 30 0.85
TC232322 weakly similar to UP|Q6T116 (Q6T116) ATPase 8 (Fragment... 30 1.5
BE020344 29 1.9
TC226825 similar to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydra... 29 2.5
TC234759 similar to UP|R30A_SCHPO (P52808) 60S ribosomal protein... 28 3.2
TC226777 similar to PIR|T01514|T01514 glycosyl transferase homol... 28 3.2
TC215241 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete 28 3.2
TC206704 similar to UP|Q94F52 (Q94F52) At2g33620/F4P9.39, partia... 28 4.2
TC215703 homologue to PIR|G96755|G96755 developmental protein ho... 28 4.2
TC217075 similar to UP|Q9SPL2 (Q9SPL2) COP1-interacting protein ... 28 5.5
BE607817 28 5.5
TC203442 weakly similar to UP|Q6PLP6 (Q6PLP6) Cell wall protein ... 27 7.2
>BG363154
Length = 509
Score = 147 bits (371), Expect = 5e-36
Identities = 82/131 (62%), Positives = 95/131 (71%), Gaps = 2/131 (1%)
Frame = +1
Query: 1 MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTK--DFPRNN 58
MELSSSF SPKP + P P L ++RN ++AVA++P + DFP N
Sbjct: 139 MELSSSFLFSPKPCS-------CPPPNYSLSPLIRNRNPNFRIVAVALEPQQQQDFPSGN 297
Query: 59 NPQRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCL 118
+PQRLLKELAERKK T+PK K PPR +IL+PP+DD KLAERFLNSPQL LK FPLLSSCL
Sbjct: 298 SPQRLLKELAERKKATSPK-KGPPRSFILRPPIDDNKLAERFLNSPQLCLKLFPLLSSCL 474
Query: 119 PSLRLNNADKL 129
PS RLNNADKL
Sbjct: 475 PSSRLNNADKL 507
>TC211144
Length = 467
Score = 145 bits (365), Expect = 2e-35
Identities = 76/113 (67%), Positives = 89/113 (78%), Gaps = 8/113 (7%)
Frame = +1
Query: 1 MELSSSFSLSPKPT-----NFSFSSSFSPFPIQILIKNTKSR---NYPLHVLAVAIDPTK 52
MELSSSF LSPKP NFSFSSS SPFPIQ+LI+N S+ P ++AVA++P +
Sbjct: 130 MELSSSFLLSPKPCSSPPPNFSFSSSLSPFPIQLLIRNGNSKFPQTPPFRIIAVALEPQQ 309
Query: 53 DFPRNNNPQRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQ 105
DFP N+PQRLLKELAERKK T+PK K PPRR+IL+PP+DD KLAERFLNSPQ
Sbjct: 310 DFPSGNSPQRLLKELAERKKATSPK-KGPPRRFILRPPIDDNKLAERFLNSPQ 465
>TC232957
Length = 430
Score = 142 bits (358), Expect = 2e-34
Identities = 80/144 (55%), Positives = 102/144 (70%)
Frame = +2
Query: 1 MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNP 60
MELSSSF+ S FSFSSS +P R L V AV IDP ++ P+N+ P
Sbjct: 35 MELSSSFTHSHNL--FSFSSSLTP----------TLRRQTLRVSAVTIDPPQELPKNS-P 175
Query: 61 QRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPS 120
QRLLKELAERK+ + ++PPRR+IL PLDDK++A+RFLNSPQL+L +FPLLSSCLP
Sbjct: 176 QRLLKELAERKRPSPRTPRAPPRRFILTRPLDDKRVADRFLNSPQLALNTFPLLSSCLPF 355
Query: 121 LRLNNADKLWIDEYLLEAKQALGY 144
L++ DK W++ +L+EAKQALGY
Sbjct: 356 SPLDHLDKSWMENHLMEAKQALGY 427
>BF425002
Length = 416
Score = 112 bits (279), Expect = 2e-25
Identities = 66/115 (57%), Positives = 77/115 (66%), Gaps = 2/115 (1%)
Frame = +3
Query: 1 MELSSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTK--DFPRNN 58
MELSSSF SP LI+N RN ++AVA++P + DFP N
Sbjct: 138 MELSSSFLFSP------------------LIRN---RNPNFRIVAVALEPQQQQDFPSGN 254
Query: 59 NPQRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPL 113
+PQRLLKELAERKK T+PK K PPRR+IL+PP+DD KLAERFLNSPQL LK FPL
Sbjct: 255 SPQRLLKELAERKKATSPK-KGPPRRFILRPPIDDNKLAERFLNSPQLCLKLFPL 416
>TC221385
Length = 622
Score = 109 bits (273), Expect = 1e-24
Identities = 50/61 (81%), Positives = 57/61 (92%)
Frame = +1
Query: 202 AEFFLQRYPRELPGPMRERVYGLIGKENLPKWIKAASLQNLIFPYDNMDKIVRRDREGPV 261
AEFFLQRYPRE PGPMRERV+GLIGK +LPKWIKAASL NLIF +D+MDKI+RR++EGPV
Sbjct: 1 AEFFLQRYPRESPGPMRERVFGLIGKRSLPKWIKAASLHNLIFTFDDMDKIMRREKEGPV 180
Query: 262 K 262
K
Sbjct: 181 K 183
>TC218482
Length = 1214
Score = 45.1 bits (105), Expect = 3e-05
Identities = 36/148 (24%), Positives = 70/148 (46%), Gaps = 3/148 (2%)
Frame = +3
Query: 113 LLSSCLPSLRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQFD--TLLYLAFQ 170
L +C+P +W + + LGY LE ++ + + A+ + L L F
Sbjct: 381 LFRACVPPGMHRFRGNIWDYDTRPHVMKTLGYPLEMADRIPEITEARNIELGLGLQLCFM 560
Query: 171 HPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGP-MRERVYGLIGKEN 229
HPS + K +H R + RL ++GQ + +L +AE L ++ + PG ++E+ L+ +
Sbjct: 561 HPS--KYKFEHPRFCYERLEYIGQKIQDLVMAERLLMKH-LDAPGLWLQEKHRHLLMNKY 731
Query: 230 LPKWIKAASLQNLIFPYDNMDKIVRRDR 257
++++A L +I D + +R
Sbjct: 732 CGRYLRAKHLHRVIIFDDKVQDTYEHNR 815
>TC204902 UP|NO51_SOYBN (P04671) Nodulin C51 precursor, complete
Length = 972
Score = 33.9 bits (76), Expect = 0.077
Identities = 20/78 (25%), Positives = 35/78 (44%)
Frame = +2
Query: 39 YPLHVLAVAIDPTKDFPRNNNPQRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAE 98
Y V + ++P++ FPR NNPQ K + ++ + PP R P L
Sbjct: 338 YQTSVTNLQVEPSEVFPRKNNPQGGRKSKLDDHQVQPLSFRLPPFRLPPMPKLGPTSPII 517
Query: 99 RFLNSPQLSLKSFPLLSS 116
R + SP ++ + L+ +
Sbjct: 518 RTIPSPPIAPRDLSLIET 571
>CF922648
Length = 570
Score = 31.6 bits (70), Expect = 0.38
Identities = 23/59 (38%), Positives = 27/59 (44%), Gaps = 3/59 (5%)
Frame = +2
Query: 50 PTKDFPRNN--NPQRLL-KELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQ 105
P ++ P N NP L K KIT SK PP+ I PP K A+R L PQ
Sbjct: 125 PPREIPPQNLKNPPPLKGKNKKAPPKITPKISKPPPKTGIQNPPKKFGKFAKRALGPPQ 301
Score = 27.7 bits (60), Expect = 5.5
Identities = 22/62 (35%), Positives = 27/62 (43%), Gaps = 7/62 (11%)
Frame = +3
Query: 76 PKSKSPPRRYILKPPLDDK------KLAERFLNSPQLSLKSFPLLSSCLPSLRLNNADK- 128
P+ KSPP+ PPL +K KL ++F N P P S P L N K
Sbjct: 126 PQGKSPPKI*KTPPPLREKIKKPPQKLPQKFPNPP-------PKRESKTPQKNLGNLPKG 284
Query: 129 LW 130
LW
Sbjct: 285 LW 290
>TC226826 similar to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydratase, small
subunit, partial (23%)
Length = 435
Score = 30.4 bits (67), Expect = 0.85
Identities = 30/104 (28%), Positives = 39/104 (36%), Gaps = 24/104 (23%)
Frame = +1
Query: 3 LSSSFSLSPKP------TNFSFSSSFSPFPI-QILIKNTKSRNYPLHVL----------- 44
L +S SL P T F +S SP P Q L ++S + P H L
Sbjct: 94 LGTSHSLPPPNSPSLTVTLFYITSFHSPLPNHQTLATGSQSLSKP-HALNPARPPPPPSM 270
Query: 45 ------AVAIDPTKDFPRNNNPQRLLKELAERKKITNPKSKSPP 82
A P + FPR+ +P L + R P S SPP
Sbjct: 271 ASATSSATTSTPIRSFPRSTSPSSLRSPTSTRSSAPTPSSASPP 402
>TC232322 weakly similar to UP|Q6T116 (Q6T116) ATPase 8 (Fragment), partial
(42%)
Length = 521
Score = 29.6 bits (65), Expect = 1.5
Identities = 37/124 (29%), Positives = 48/124 (37%), Gaps = 4/124 (3%)
Frame = +3
Query: 1 MELSSSFSL---SPK-PTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPR 56
M LS S SL SP P F +S SF +K K R VL + +D R
Sbjct: 108 MLLSLSLSLPLISPHVPLLFCYSCSFG-------VKTRKKRTERNFVLGTKRNQWEDVQR 266
Query: 57 NNNPQRLLKELAERKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSS 116
+ R + R NP S P R LKP K + L +L S LL++
Sbjct: 267 SRRSPRTIPMECPRHPNQNPNLNSNPTRTPLKPKHPPPKW-NKLLLLTTTTLMSTKLLTT 443
Query: 117 CLPS 120
P+
Sbjct: 444 TKPT 455
>BE020344
Length = 354
Score = 29.3 bits (64), Expect = 1.9
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 1/81 (1%)
Frame = +2
Query: 19 SSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELA-ERKKITNPK 77
+S SP QI IK S + DP+K FP N + +R L +L+ E K+ N +
Sbjct: 95 ASKMSPDDSQIQIKVPSSNSDEQ-----LPDPSKSFPGNEDLERSLHKLSKELKETQNER 259
Query: 78 SKSPPRRYILKPPLDDKKLAE 98
+K+ LK L +K+ E
Sbjct: 260 NKAVQELTRLKQHLLEKESEE 322
>TC226825 similar to UP|Q9ZW85 (Q9ZW85) 3-isopropylmalate dehydratase, small
subunit, partial (71%)
Length = 1121
Score = 28.9 bits (63), Expect = 2.5
Identities = 12/33 (36%), Positives = 16/33 (48%)
Frame = +3
Query: 50 PTKDFPRNNNPQRLLKELAERKKITNPKSKSPP 82
PT+ FP + +P L + R P S SPP
Sbjct: 255 PTRSFPPSTSPSSLPSPTSTRSSAPTPSSASPP 353
>TC234759 similar to UP|R30A_SCHPO (P52808) 60S ribosomal protein L30-1
(L32), partial (93%)
Length = 681
Score = 28.5 bits (62), Expect = 3.2
Identities = 25/87 (28%), Positives = 43/87 (48%), Gaps = 9/87 (10%)
Frame = +1
Query: 53 DFPRNNNPQRLLKELAERKKITNPKSKSPPRR-------YILKPPLDDKKLAERFLNSP- 104
D P ++NP +L+ ++ +IT +S+ PPRR L P ++ ++ SP
Sbjct: 7 DRPPSDNPSPILRPDPDQDQITTNQSRWPPRRRPRPVTPSTLASPW**SRVRSFWVTSPL 186
Query: 105 -QLSLKSFPLLSSCLPSLRLNNADKLW 130
+LS + P SS P+ L +A + W
Sbjct: 187 *RLSALARPSWSSS-PATPLLSASRSW 264
>TC226777 similar to PIR|T01514|T01514 glycosyl transferase homolog T10M13.14
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(83%)
Length = 1247
Score = 28.5 bits (62), Expect = 3.2
Identities = 38/145 (26%), Positives = 55/145 (37%)
Frame = +1
Query: 13 PTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELAERKK 72
P+ ++S+S +P P NT S + + PT + P +N P R R
Sbjct: 172 PSPSTWSTSAAPSPPSTPSSNTPSARRTSSSTSSSPKPTSN-PSSNPPSR---N*TSRSI 339
Query: 73 ITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSLRLNNADKLWID 132
P+S + PPL DK N+P + + L SS S L +W+
Sbjct: 340 TLIPRSSAT*-----SPPLSDKPS-----NNPSTTPEIISLTSSNPVSKELYT*IPIWL* 489
Query: 133 EYLLEAKQALGYSLEPSETLEDDNP 157
L + A LEPS L P
Sbjct: 490 STTLPSSGAPA*VLEPSAPLNTATP 564
>TC215241 homologue to UP|Q6WV72 (Q6WV72) Histone H4, complete
Length = 707
Score = 28.5 bits (62), Expect = 3.2
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = -3
Query: 22 FSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFP 55
FSPF IK K + P+H V +D +K FP
Sbjct: 582 FSPFTTNTTIKKNKKKTTPIHASFVFLD-SKTFP 484
>TC206704 similar to UP|Q94F52 (Q94F52) At2g33620/F4P9.39, partial (28%)
Length = 1556
Score = 28.1 bits (61), Expect = 4.2
Identities = 13/24 (54%), Positives = 15/24 (62%)
Frame = -3
Query: 164 LLYLAFQHPSCERTKQKHVRSGHS 187
LL L F HP+ E HVRSGH+
Sbjct: 168 LLLLGFVHPNRELLPPCHVRSGHA 97
>TC215703 homologue to PIR|G96755|G96755 developmental protein homolog DG1118
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , complete
Length = 896
Score = 28.1 bits (61), Expect = 4.2
Identities = 23/66 (34%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Frame = -2
Query: 194 QYVLELALAEFFLQRYPRELPGPMRERVYGLIGKENLPKWIKAASLQNLIFP-YDNMDKI 252
+++L LAL LQR+ REL + V+ L+ +P + A L+ +FP + N D
Sbjct: 190 RFLLLLALTRLPLQRFRREL--QFHDLVHQLL---RVPHPYRFALLRVFLFPKFRNTDIE 26
Query: 253 VRRDRE 258
RDRE
Sbjct: 25 TERDRE 8
>TC217075 similar to UP|Q9SPL2 (Q9SPL2) COP1-interacting protein CIP8
(At5g64920), partial (34%)
Length = 1267
Score = 27.7 bits (60), Expect = 5.5
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = -2
Query: 5 SSFSLSPKPTNFSFSSSFSPFPIQILIKN 33
SS S SP ++FSFSSS S P IL K+
Sbjct: 414 SSNSSSPLSSSFSFSSSLSQPPSSILRKS 328
>BE607817
Length = 463
Score = 27.7 bits (60), Expect = 5.5
Identities = 15/43 (34%), Positives = 20/43 (45%)
Frame = -2
Query: 41 LHVLAVAIDPTKDFPRNNNPQRLLKELAERKKITNPKSKSPPR 83
+HVL + D +FPR +NP NPK K PP+
Sbjct: 210 VHVLLMMYDG*SNFPRKSNP------------ALNPKIKQPPK 118
>TC203442 weakly similar to UP|Q6PLP6 (Q6PLP6) Cell wall protein GP2
(Fragment), partial (6%)
Length = 980
Score = 27.3 bits (59), Expect = 7.2
Identities = 19/44 (43%), Positives = 22/44 (49%)
Frame = +3
Query: 4 SSSFSLSPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVA 47
SS+ L P PTN SFSS F P T + PL V A+A
Sbjct: 177 SSAPVLRPIPTNRSFSSPFRSLP-------TPTTRKPLTVFAMA 287
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,878,638
Number of Sequences: 63676
Number of extensions: 209742
Number of successful extensions: 1453
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 1433
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1443
length of query: 262
length of database: 12,639,632
effective HSP length: 95
effective length of query: 167
effective length of database: 6,590,412
effective search space: 1100598804
effective search space used: 1100598804
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146561.3


