

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
(519 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
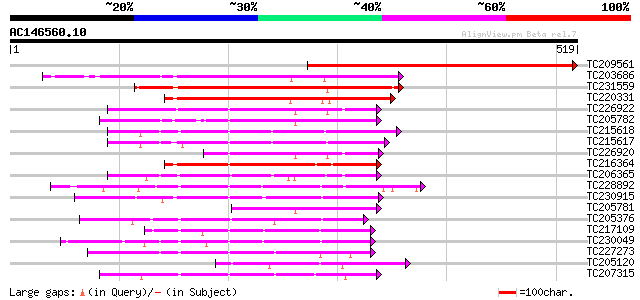
Score E
Sequences producing significant alignments: (bits) Value
TC209561 similar to UP|Q40542 (Q40542) NPK2 , partial (48%) 467 e-132
TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete 238 5e-63
TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%) 226 1e-59
TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partia... 221 8e-58
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 170 1e-42
TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 155 3e-38
TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 149 3e-36
TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 146 2e-35
TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial... 135 4e-32
TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%) 128 5e-30
TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 120 2e-27
TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein ... 101 7e-22
TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-sp... 101 7e-22
TC205781 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 95 9e-20
TC205376 similar to UP|Q9MB45 (Q9MB45) IRE homolog 1 (Fragment),... 94 1e-19
TC217109 UP|Q39886 (Q39886) Protein kinase , complete 94 2e-19
TC230049 similar to UP|Q9MAV2 (Q9MAV2) F24O1.13, partial (47%) 94 2e-19
TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable serin... 93 2e-19
TC205120 weakly similar to UP|Q7KSD3 (Q7KSD3) CG7693-PA (Cg7693-... 92 6e-19
TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kina... 91 9e-19
>TC209561 similar to UP|Q40542 (Q40542) NPK2 , partial (48%)
Length = 869
Score = 467 bits (1201), Expect = e-132
Identities = 222/247 (89%), Positives = 233/247 (93%)
Frame = +2
Query: 273 LLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAE 332
L E GTGEFPYTANEGPVNLMLQILDDPSPSP K KFSPEFCSFVDACLQKDPD RPTAE
Sbjct: 2 LFECGTGEFPYTANEGPVNLMLQILDDPSPSPLKNKFSPEFCSFVDACLQKDPDTRPTAE 181
Query: 333 QLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLADMLTIHYYLLFDGPDDSWQHTRNF 392
QLL HPFITK++ AKVDL GFVRSVFDPTQR+KDLADMLTIHYYLLFDGPDD WQHT+N
Sbjct: 182 QLLSHPFITKHDDAKVDLAGFVRSVFDPTQRLKDLADMLTIHYYLLFDGPDDLWQHTKNL 361
Query: 393 YNENSIFSFSGKQHIGPNNIFTTLSSIRTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIR 452
Y+E+SIFSFSGKQH GP+NIFT+LSSIRTTL+GDWPPEKLVHVVE+LQCRAHGEDGVAIR
Sbjct: 362 YSESSIFSFSGKQHCGPSNIFTSLSSIRTTLVGDWPPEKLVHVVERLQCRAHGEDGVAIR 541
Query: 453 VSGSFIIGNQFLICGDGVQVEGLPNFKDLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVK 512
VSGSFIIGNQFLICGDG+QVEGLPNFKDLGIDI SKRMGTFHEQFIVEP S IGCYTIVK
Sbjct: 542 VSGSFIIGNQFLICGDGIQVEGLPNFKDLGIDIPSKRMGTFHEQFIVEPKSLIGCYTIVK 721
Query: 513 QELYINQ 519
QELYINQ
Sbjct: 722 QELYINQ 742
>TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete
Length = 1502
Score = 238 bits (607), Expect = 5e-63
Identities = 139/339 (41%), Positives = 203/339 (59%), Gaps = 9/339 (2%)
Frame = +2
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV +++ ++ + D N+ T ++ + +
Sbjct: 170 ESGTFKDG---DLLVNRDGVRIVSQNDVEAPPPIKPTD----NQLTLA----DIDVIKVV 316
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G VVQ H T + ALK I + E+ R+Q+ E++ +A C +V + +F
Sbjct: 317 GKGNGGVVQLVQHKWTSQFFALKVIQMNIEESMRKQIAQELKINQQAQC-PYVVVCYQSF 493
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
Y ++G ISI LEYMDGGSLAD+L+ +TIPE L+++ +++L+GL YLH ++++HRD+
Sbjct: 494 Y--ENGVISIILEYMDGGSLADLLKKVKTIPEDYLAAICKQVLKGLVYLHHEKHIIHRDL 667
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIR--NESYSYPADIW 267
KP+NLL+N GE KITDFG+SA +E++ TF+GT YMSPERI Y+Y +DIW
Sbjct: 668 KPSNLLINHIGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNYKSDIW 847
Query: 268 SLGLALLESGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACL 321
SLGL LLE G FPY E L+ I+D P P P E+FS EFCSF+ ACL
Sbjct: 848 SLGLILLECALGRFPYAPPDQSETWESIFELIETIVDKPPPIPPSEQFSTEFCSFISACL 1027
Query: 322 QKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
QKDP +R +A++L+ HPF+ Y+ +VDL + + P
Sbjct: 1028QKDPKDRLSAQELMAHPFVNMYDDLEVDLSAYFSNAGSP 1144
>TC231559 homologue to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (71%)
Length = 867
Score = 226 bits (577), Expect = 1e-59
Identities = 118/252 (46%), Positives = 170/252 (66%), Gaps = 6/252 (2%)
Frame = +2
Query: 115 INIFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILR 174
+NI +++ R+Q++ E++ + C +V +H ++ +G IS+ LEYMD GSLAD+++
Sbjct: 11 VNI-QEDIRKQIVQELKINQASQCPHVVVCYHSFYH---NGVISLVLEYMDRGSLADVIK 178
Query: 175 MHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLE 234
+TI EP L+ +F+++L+GL YLH R+++HRDIKP+NLLVN KGE KITDFG+SA L
Sbjct: 179 QVKTILEPYLAVVFKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLA 358
Query: 235 NSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP----- 289
+S+ TFVGT YMSPERI +Y Y +DIWSLG+ +LE G FPY +E
Sbjct: 359 SSMGQRDTFVGTYNYMSPERISGSTYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQSWPS 538
Query: 290 -VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKV 348
L+ I++ P PS ++FSPEFCSFV +C+QKDP +R T+ +LL HPFI K+E +
Sbjct: 539 FYELLAAIVESPPPSAPPDQFSPEFCSFVSSCIQKDPRDRLTSLKLLDHPFIKKFEDKDL 718
Query: 349 DLPGFVRSVFDP 360
DL G + +P
Sbjct: 719 DL-GILAGSLEP 751
>TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (62%)
Length = 669
Score = 221 bits (562), Expect = 8e-58
Identities = 113/219 (51%), Positives = 153/219 (69%), Gaps = 7/219 (3%)
Frame = +1
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
+V + +FY +G ISI LEYMDGGSL D+L +TIPE LS++ +++L+GL YLH
Sbjct: 13 VVVCYNSFY--HNGVISIILEYMDGGSLEDLLSKVKTIPESYLSAICKQVLKGLMYLHYA 186
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERI--RNES 259
++++HRD+KP+NLL+N +GE KITDFG+S +EN+ TF+GT +YMSPERI
Sbjct: 187 KHIIHRDLKPSNLLINHRGEVKITDFGVSVIMENTSGQANTFIGTYSYMSPERIIGNQHG 366
Query: 260 YSYPADIWSLGLALLESGTGEFPYTA--NEGPVN---LMLQILDDPSPSPSKEKFSPEFC 314
Y+Y +DIWSLGL LL+ TG+FPYT EG N L+ I++ PSPS + FSPEFC
Sbjct: 367 YNYKSDIWSLGLILLKCATGQFPYTPPDREGWENIFQLIEVIVEKPSPSAPSDDFSPEFC 546
Query: 315 SFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGF 353
SF+ ACLQK+P +RP+A L+ HPFI +E VDL +
Sbjct: 547 SFISACLQKNPGDRPSARDLINHPFINMHEDLNVDLSAY 663
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 170 bits (431), Expect = 1e-42
Identities = 108/261 (41%), Positives = 145/261 (55%), Gaps = 10/261 (3%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGA 148
+G G V + H T ALK I+ + +R++ L+E L +V FH +
Sbjct: 235 LGHGNGGTVYKVRHKATSATYALKIIHSDTDATRRRRALSETSILRRVTDCPHVVRFHSS 414
Query: 149 FYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRD 208
F P SG ++I +EYMDGG+L L T E L+ + + +L GL+YLH R + HRD
Sbjct: 415 FEKP-SGDVAILMEYMDGGTLETALAASGTFSEERLAKVARDVLEGLAYLHA-RNIAHRD 588
Query: 209 IKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY-----SYP 263
IKPAN+LVN +G+ KI DFG+S + S+ C ++VGT YMSP+R E+Y +
Sbjct: 589 IKPANILVNSEGDVKIADFGVSKLMCRSLEACNSYVGTCAYMSPDRFDPEAYGGNYNGFA 768
Query: 264 ADIWSLGLALLESGTGEFPY-TANEGP--VNLMLQI-LDDPSPSPSKEKFSPEFCSFVDA 319
ADIWSLGL L E G FP+ A + P LM I DP P E SPEF FV+
Sbjct: 769 ADIWSLGLTLFELYVGHFPFLQAGQRPDWATLMCAICFGDPPSLP--ETASPEFRDFVEC 942
Query: 320 CLQKDPDNRPTAEQLLLHPFI 340
CL+K+ R T QLL HPF+
Sbjct: 943 CLKKESGERWTTAQLLTHPFV 1005
>TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (91%)
Length = 1377
Score = 155 bits (393), Expect = 3e-38
Identities = 100/265 (37%), Positives = 149/265 (55%), Gaps = 7/265 (2%)
Frame = +2
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKI-NIFEKEKRQQLLTEIRTLCEAPCYEG 141
E+ IGSG+ V + +H + RV ALK I E+ R+Q+ EI+ L +
Sbjct: 278 ELERLNRIGSGSGGTVYKVVHRTSGRVYALKVIYGHHEESVRRQIHREIQILRDVDD-AN 454
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
+V+ H + + +I + LE+MDGGSL H T E L+ + +++LRGL+YLH
Sbjct: 455 VVKCHEMY--DQNSEIQVLLEFMDGGSLEG---KHIT-QEQQLADLSRQILRGLAYLHR- 613
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY- 260
R++VHRDIKP+NLL+N + + KI DFG+ L ++ C + VGT+ YMSPERI +
Sbjct: 614 RHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDIND 793
Query: 261 ----SYPADIWSLGLALLESGTGEFPY-TANEGPVNLMLQILDDPSPSPSKEKFSPEFCS 315
+Y DIWS G+++LE G FP+ +G ++ + P + SP F
Sbjct: 794 GQYDAYAGDIWSFGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPPSASPHFKD 973
Query: 316 FVDACLQKDPDNRPTAEQLLLHPFI 340
F+ CLQ+DP R +A +LL HPFI
Sbjct: 974 FILRCLQRDPSRRWSASRLLEHPFI 1048
>TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (73%)
Length = 2291
Score = 149 bits (376), Expect = 3e-36
Identities = 94/275 (34%), Positives = 150/275 (54%), Gaps = 6/275 (2%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIF-----EKEKRQQLLTEIRTLCEAPCYEGLVE 144
+G G V + ++ A+K++ + KE +QL EI L + + +V+
Sbjct: 634 LGRGTFGHVYLGFNSENGQMCAIKEVKVVFDDHTSKECLKQLNQEINLLNQLS-HPNIVQ 810
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
+HG+ +S +S+ LEY+ GGS+ +L+ + EP++ + ++++ GL+YLHG R
Sbjct: 811 YHGSELVEES--LSVYLEYVSGGSIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHG-RNT 981
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN-ESYSYP 263
VHRDIK AN+LV+ GE K+ DFG++ + +S +M +F G+ +M+PE + N YS P
Sbjct: 982 VHRDIKGANILVDPNGEIKLADFGMAKHINSSASM-LSFKGSPYWMAPEVVMNTNGYSLP 1158
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
DIWSLG ++E T + P+ EG V + +I + E S + F+ CLQ+
Sbjct: 1159VDIWSLGCTIIEMATSKPPWNQYEG-VAAIFKIGNSKDMPEIPEHLSNDAKKFIKLCLQR 1335
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVF 358
DP RPTA++LL HPFI K R F
Sbjct: 1336DPLARPTAQKLLDHPFIRDQSATKAANVSITRDAF 1440
>TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (51%)
Length = 1451
Score = 146 bits (368), Expect = 2e-35
Identities = 91/266 (34%), Positives = 150/266 (56%), Gaps = 8/266 (3%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIF-----EKEKRQQLLTEIRTLCEAPCYEGLVE 144
+G G V + + ++ A+K++ + KE +QL EI L + + +V+
Sbjct: 672 LGRGTFGHVYLGFNSDSGQLCAIKEVRVVCDDQSSKECLKQLNQEIHLLSQLS-HPNIVQ 848
Query: 145 FHGAFYTPDSGQ--ISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVR 202
++G+ D G+ +S+ LEY+ GGS+ +L+ + EP++ + ++++ GLSYLHG R
Sbjct: 849 YYGS----DLGEETLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLSYLHG-R 1013
Query: 203 YLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN-ESYS 261
VHRDIK AN+LV+ GE K+ DFG++ + +S +M +F G+ +M+PE + N YS
Sbjct: 1014NTVHRDIKGANILVDPNGEIKLADFGMAKHINSSSSM-LSFKGSPYWMAPEVVMNTNGYS 1190
Query: 262 YPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACL 321
P DIWSLG +LE T + P+ EG + +I + + S + +F+ CL
Sbjct: 1191LPVDIWSLGCTILEMATSKPPWNQYEGAA-AIFKIGNSRDMPEIPDHLSSDAKNFIQLCL 1367
Query: 322 QKDPDNRPTAEQLLLHPFITKYETAK 347
Q+DP RPTA++L+ HPFI K
Sbjct: 1368QRDPSARPTAQKLIEHPFIRDQSATK 1445
>TC226920 weakly similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (49%)
Length = 722
Score = 135 bits (340), Expect = 4e-32
Identities = 81/174 (46%), Positives = 104/174 (59%), Gaps = 9/174 (5%)
Frame = +2
Query: 178 TIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSV 237
T E L+ + + +L GL+YLH R + HRDIKPAN+LVN +GE KI DFG+S + ++
Sbjct: 41 TFSEERLAKVARDVLEGLAYLHA-RNIAHRDIKPANILVNSEGEVKIADFGVSKLMCRTL 217
Query: 238 AMCATFVGTVTYMSPERIRNESY-----SYPADIWSLGLALLESGTGEFPY-TANEGP-- 289
C ++VGT YMSP+R E+Y + ADIWSLGL L E G FP+ A + P
Sbjct: 218 EACNSYVGTCAYMSPDRFDPEAYGGNYNGFAADIWSLGLTLFELYVGHFPFLQAGQRPDW 397
Query: 290 VNLMLQI-LDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITK 342
LM I DP P E SPEF FV+ CL+K+ R TA QLL HPF+ K
Sbjct: 398 ATLMCAICFSDPPSLP--ETASPEFHDFVECCLKKESGERWTAAQLLTHPFVCK 553
>TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%)
Length = 2224
Score = 128 bits (322), Expect = 5e-30
Identities = 74/200 (37%), Positives = 121/200 (60%), Gaps = 1/200 (0%)
Frame = +1
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
+ E++G++ + ++ I +EYM GGS+AD+++ + E ++ + + LL + YLH
Sbjct: 34 ITEYYGSYL--NQTKLWIIMEYMAGGSVADLIQSGPPLDEMSIACILRDLLHAVDYLHS- 204
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRN-ESY 260
+HRDIK AN+L++ G+ K+ DFG+SA L +++ TFVGT +M+PE I+N + Y
Sbjct: 205 EGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQNTDGY 384
Query: 261 SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDAC 320
+ ADIWSLG+ +E GE P A+ P+ ++ I+ +P + FS FV C
Sbjct: 385 NEKADIWSLGITAIEMAKGE-PPLADLHPMRVLF-IIPRENPPQLDDHFSRPLKEFVSLC 558
Query: 321 LQKDPDNRPTAEQLLLHPFI 340
L+K P RP+A++LL FI
Sbjct: 559 LKKVPAERPSAKELLKDRFI 618
>TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (37%)
Length = 1866
Score = 120 bits (300), Expect = 2e-27
Identities = 89/265 (33%), Positives = 143/265 (53%), Gaps = 14/265 (5%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR-----QQLLTEIRTLCEAPCYEGLVE 144
IG G+ V A ++ T ALK++++F + + +QL EIR L + + +V+
Sbjct: 454 IGRGSYGSVYHATNLETGASCALKEVDLFPDDPKSADCIKQLEQEIRILRQLH-HPNIVQ 630
Query: 145 FHGAFYTPDSGQISIALEYMDGGSLADILRMH-RTIPEPILSSMFQKLLRGLSYLHGVRY 203
++G+ D ++ I +EY+ GSL + H + E ++ + + +L GL+YLHG +
Sbjct: 631 YYGSEIVGD--RLYIYMEYVHPGSLHKFMHEHCGAMTESVVRNFTRHILSGLAYLHGTK- 801
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGISAGL-ENSVAMCATFVGTVTYMSPE----RIRNE 258
+HRDIK ANLLV+ G K+ DFG+S L E S + + G+ +M+PE I+ E
Sbjct: 802 TIHRDIKGANLLVDASGSVKLADFGVSKILTEKSYEL--SLKGSPYWMAPELMKAAIKKE 975
Query: 259 S---YSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCS 315
S + DIWSLG ++E TG+ P++ EGP M ++L P E S E
Sbjct: 976 SSPDIAMAIDIWSLGCTIIEMLTGKPPWSEFEGP-QAMFKVLHKSPDLP--ESLSSEGQD 1146
Query: 316 FVDACLQKDPDNRPTAEQLLLHPFI 340
F+ C +++P RP+A LL H F+
Sbjct: 1147FLQQCFRRNPAERPSAAVLLTHAFV 1221
>TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein kinase,
complete
Length = 1931
Score = 101 bits (252), Expect = 7e-22
Identities = 101/373 (27%), Positives = 163/373 (43%), Gaps = 30/373 (8%)
Frame = +1
Query: 38 GGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEM-----RIFGAIGS 92
G +LL S+ +++ N T RS+ + D + GS +M ++ +G
Sbjct: 1 GTRCSLLPFSFSIHHSNP------RTLRSITKMDRSTGRGGGGSVDMFLRNYKLGKTLGI 162
Query: 93 GASSVVQRAMHIPTHRVIALKKIN---IFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G+ V+ A H+ T +A+K +N I E +++ EI+ L + ++ +
Sbjct: 163 GSFGKVKIAEHVRTGHKVAIKILNRHKIKNMEMEEKVRREIKIL-RLFMHHHIIRLYEVV 339
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
TP I + +EY+ G L D + + E FQ+++ G+ Y H +VHRD+
Sbjct: 340 ETPTD--IYVVMEYVKSGELFDYIVEKGRLQEDEARHFFQQIISGVEYCHR-NMVVHRDL 510
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYP-ADIWS 268
KP NLL++ K KI DFG+S + T G+ Y +PE I + Y+ P D+WS
Sbjct: 511 KPENLLLDSKFNIKIADFGLS-NIMRDGHFLKTSCGSPNYAAPEVISGKLYAGPEVDVWS 687
Query: 269 LGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNR 328
G+ L G P+ +E NL +I PS SP + L DP R
Sbjct: 688 CGVILYALLCGTLPFD-DENIPNLFKKIKGGIYTLPS--HLSPGARDLIPRMLVVDPMKR 858
Query: 329 PTAEQLLLHPFI--------------TKYETAKVD---LPGFVRSVFDPTQRMKDLADML 371
T ++ HP+ T + K+D L V FD Q ++ L++ +
Sbjct: 859 MTIPEIRQHPWFQVHLPRYLAVPPPDTLQQAKKIDEEILQEVVNMGFDRNQLVESLSNRI 1038
Query: 372 ----TIHYYLLFD 380
T+ YYLL D
Sbjct: 1039QNEGTVTYYLLLD 1077
>TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-specific
protein kinase homolog T16I18.40 - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (90%)
Length = 1174
Score = 101 bits (252), Expect = 7e-22
Identities = 82/287 (28%), Positives = 137/287 (47%), Gaps = 4/287 (1%)
Frame = +3
Query: 60 KCSTSRSVDESDDNEKTYRCGSHE-MRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIF 118
+ ST + + E + R S E I +G G V A + + V+ALK I
Sbjct: 3 RISTKMASQNPAEEENSKRHWSLEDFEIGKPLGRGKFGRVYVAREVKSKFVVALKVI-FK 179
Query: 119 EKEKRQQLLTEIRTLCEAPC---YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRM 175
E+ + ++ ++R E + ++ +G F+ D+ ++ + LEY G L LR
Sbjct: 180 EQIDKYRVHHQLRREMEIQTSLRHANILRLYGGFH--DADRVFLILEYAHKGELNKELRK 353
Query: 176 HRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLEN 235
+ E ++ L + L+Y H ++++HRDIKP NLL + +G KI DFG S
Sbjct: 354 KGHLTEKQAATYIVSLTKALAYCHE-KHVIHRDIKPENLLPDHEGRLKIADFGWSV---Q 521
Query: 236 SVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQ 295
S + T GT+ Y++PE N+++ Y D W+LG+ ES G P+ A E + +
Sbjct: 522 SRSKRHTMCGTLDYLAPEMAENKAHDYAVDNWTLGIRCYESLYGAPPFEA-ESQSDTFKR 698
Query: 296 ILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITK 342
I+ PS S E + + L KD R + ++++ HP+I K
Sbjct: 699 IMKVDLSFPSTPSVSIEAKNLISRLLVKDSSRRLSLQKIMEHPWIIK 839
>TC205781 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (47%)
Length = 935
Score = 94.7 bits (234), Expect = 9e-20
Identities = 53/143 (37%), Positives = 75/143 (52%), Gaps = 6/143 (4%)
Frame = +2
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY--- 260
+VHRDIKP N + + KI FG+ L ++ C VGT+ MSPERI +
Sbjct: 47 IVHRDIKPXNXXXXSRKQGKIAXFGVGRILNQTMDPCNXSVGTIANMSPERINTDINDGQ 226
Query: 261 --SYPADIWSLGLALLESGTGEFPY-TANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFV 317
+Y DIWS G+++LE G FP+ +G ++ + P + SP F F+
Sbjct: 227 YDAYAGDIWSFGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQPPEAPPSASPHFKDFI 406
Query: 318 DACLQKDPDNRPTAEQLLLHPFI 340
CLQ+DP R +A +LL HPFI
Sbjct: 407 LRCLQRDPSRRWSASRLLEHPFI 475
>TC205376 similar to UP|Q9MB45 (Q9MB45) IRE homolog 1 (Fragment), partial
(45%)
Length = 2377
Score = 94.0 bits (232), Expect = 1e-19
Identities = 85/299 (28%), Positives = 133/299 (44%), Gaps = 35/299 (11%)
Frame = +2
Query: 65 RSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIA---LKKINIFEKE 121
RS+ S + R + I I GA V A T + A LKK ++ K
Sbjct: 230 RSLRTSPIHSSRDRTSIDDFEIIKPISRGAFGRVFLAKKRTTGDLFAIKVLKKADMIRKN 409
Query: 122 KRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPE 181
+ +L E R + +V F +F ++ + + +EY++GG L +LR + E
Sbjct: 410 AVESILAE-RDILITVRNPFVVRFFYSFTCREN--LYLVMEYLNGGDLYSLLRNLGCLDE 580
Query: 182 PILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGIS-AGLENSVAMC 240
+ +++ L YLH +R +VHRD+KP NLL+ G K+TDFG+S GL NS
Sbjct: 581 EVARVYIAEVVLALEYLHSLR-VVHRDLKPDNLLIAHDGHIKLTDFGLSKVGLINSTDDL 757
Query: 241 A------------------------------TFVGTVTYMSPERIRNESYSYPADIWSLG 270
+ + VGT Y++PE + + + AD WS+G
Sbjct: 758 SGPAVNGTSLLEEDETDVFTSADQRERREKRSAVGTPDYLAPEILLGTGHGFTADWWSVG 937
Query: 271 LALLESGTGEFPYTANEGPVNLMLQILDDPSPSPS-KEKFSPEFCSFVDACLQKDPDNR 328
+ L E G P+ A E P + IL+ P P+ E+ SP+ +D L +DP+ R
Sbjct: 938 VILFELLVGIPPFNA-EHPQTIFDNILNRKIPWPAVPEEMSPQAQDLIDRLLTEDPNQR 1111
>TC217109 UP|Q39886 (Q39886) Protein kinase , complete
Length = 1940
Score = 93.6 bits (231), Expect = 2e-19
Identities = 65/214 (30%), Positives = 104/214 (48%), Gaps = 2/214 (0%)
Frame = +1
Query: 124 QQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRM--HRTIPE 181
+Q + E+ TL ++ +++F A P I EY+ GSL L H+T+
Sbjct: 937 KQFIREV-TLLSRLHHQNVIKFSAACRKPPV--YCIITEYLAEGSLRAYLHKLEHQTVSL 1107
Query: 182 PILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCA 241
L + + RG+ Y+H + ++HRD+KP N+L+N KI DFGI+ E S + A
Sbjct: 1108 QKLIAFALDIARGMEYIHS-QGVIHRDLKPENVLINEDNHLKIADFGIACE-EASCDLLA 1281
Query: 242 TFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPS 301
GT +M+PE I+ +SY D++S GL L E TG PY + P+ +++ S
Sbjct: 1282 DDPGTYRWMAPEMIKRKSYGKKVDVYSFGLILWEMLTGTIPY-EDMNPIQAAFAVVNKNS 1458
Query: 302 PSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLL 335
P + ++ C PD RP Q++
Sbjct: 1459 RPIIPSNCPPAMRALIEQCWSLQPDKRPEFWQVV 1560
>TC230049 similar to UP|Q9MAV2 (Q9MAV2) F24O1.13, partial (47%)
Length = 1216
Score = 93.6 bits (231), Expect = 2e-19
Identities = 76/297 (25%), Positives = 134/297 (44%), Gaps = 8/297 (2%)
Frame = +2
Query: 47 SYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPT 106
S G YN + K S + + E+ + ++ I SG S + R ++
Sbjct: 56 SLGEYN-RAVSWSKYLVSPGAEIKGEGEEEWSADMSQLLIGSKFASGRHSRIYRGVY--K 226
Query: 107 HRVIALKKINIFEKEK------RQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIA 160
+ +A+K I+ E+++ +Q +E+ L + ++ F A P I
Sbjct: 227 QKDVAIKLISQPEEDEDLAAFLEKQFTSEVSLLLRLG-HPNIITFIAACKKPPV--FCII 397
Query: 161 LEYMDGGSLADILRMHRT--IPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNL 218
EY+ GGSL L + +P ++ + + RG+ YLH + ++HRD+K NLL+
Sbjct: 398 TEYLAGGSLGKFLHHQQPNILPLKLVLKLALDIARGMKYLHS-QGILHRDLKSENLLLGE 574
Query: 219 KGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGT 278
K+ DFGIS LE+ F GT +M+PE I+ + ++ D++S G+ L E T
Sbjct: 575 DMCVKVADFGISC-LESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLT 751
Query: 279 GEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLL 335
G+ P+ N P + + P K F ++ C +PD RP ++++
Sbjct: 752 GKTPFD-NMTPEQAAYAVSHKNARPPLPSKCPWAFSDLINRCWSSNPDKRPHFDEIV 919
>TC227273 weakly similar to UP|KGS9_YEAST (P43637) Probable
serine/threonine-protein kinase YGL179C , partial (8%)
Length = 2373
Score = 93.2 bits (230), Expect = 2e-19
Identities = 74/292 (25%), Positives = 131/292 (44%), Gaps = 28/292 (9%)
Frame = +3
Query: 72 DNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIR 131
D + +++++ +GSG+ + R + I + K + ++ E+
Sbjct: 975 DGADVWEIDTNQLKYENKVGSGSFGDLYRGTYCSQDVAIKVLKPERISTDMLREFAQEVY 1154
Query: 132 TLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPE-PILSSMFQK 190
+ + ++ +V+F GA P + + I E+M GSL D L R + + P L +
Sbjct: 1155 IMRKIR-HKNVVQFIGACTRPPN--LCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVAID 1325
Query: 191 LLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYM 250
+ +G++YLH ++HRD+K ANLL++ K+ DFG++ S M A GT +M
Sbjct: 1326 VSKGMNYLHQ-NNIIHRDLKTANLLMDENEVVKVADFGVARVQTQSGVMTAE-TGTYRWM 1499
Query: 251 SPERIRNESYSYPADIWSLGLALLESGTGEFPY---TANEGPVNLMLQILDDPSPSPSKE 307
+PE I ++ Y AD++S G+AL E TGE PY T + V ++ + L P +
Sbjct: 1500 APEVIEHKPYDQKADVFSFGIALWELLTGELPYSCLTPLQAAVGVVQKGLRPTIPKNTHP 1679
Query: 308 KFS------------------------PEFCSFVDACLQKDPDNRPTAEQLL 335
+ S P + C Q+DP RP +++
Sbjct: 1680 RLS*LHYKQQLAWCRRDQLSTIPKNTHPRLSELLQRCWQQDPTQRPNFSEVI 1835
>TC205120 weakly similar to UP|Q7KSD3 (Q7KSD3) CG7693-PA (Cg7693-pb)
(RE53265p), partial (20%)
Length = 2170
Score = 92.0 bits (227), Expect = 6e-19
Identities = 58/186 (31%), Positives = 97/186 (51%), Gaps = 7/186 (3%)
Frame = +2
Query: 189 QKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENS---VAMCATFVG 245
++ L+ L YLH + +HRD+K N+L++ G+ K+ DFG+SA + ++ TFVG
Sbjct: 2 KETLKALEYLHRHGH-IHRDVKAGNILLDDNGQVKLADFGVSACMFDTGDRQRSRNTFVG 178
Query: 246 TVTYMSPERIR-NESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPS- 303
T +++PE ++ Y++ ADIWS G+ LE G P+ + P+ ++L + + P
Sbjct: 179 TPCWIAPEVLQPGTGYNFKADIWSFGITALELAHGHAPF-SKYPPMKVLLMTIQNAPPGL 355
Query: 304 --PSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPT 361
KFS F V CL KD RP+ E+LL H F + + ++ + +
Sbjct: 356 DYDRDRKFSKSFKEMVAMCLVKDQTKRPSVEKLLKHSFFKQAKPPELSVKKLFADLPPLW 535
Query: 362 QRMKDL 367
R+K L
Sbjct: 536 NRVKSL 553
>TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1063
Score = 91.3 bits (225), Expect = 9e-19
Identities = 74/262 (28%), Positives = 117/262 (44%), Gaps = 4/262 (1%)
Frame = +3
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFE--KEKRQQLLTEIRTLCEAPCYE 140
E ++ +G G V R H +++ A K I E R+ + E + + +
Sbjct: 54 EYQVLEELGRGRFGTVFRCFHRTSNKFYAAKLIEKRRLLNEDRRCIEMEAKAMSFLSPHP 233
Query: 141 GLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHG 200
+++ AF DS SI LE +L D + + EP +S+ ++LL +++ H
Sbjct: 234 NILQIMDAFEDADS--CSIVLELCQPHTLLDRIAAQGPLTEPHAASLLKQLLEAVAHCHA 407
Query: 201 VRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY 260
+ L HRDIKP N+L + + K++DFG + L +M VGT Y++PE I Y
Sbjct: 408 -QGLAHRDIKPENILFDEGNKLKLSDFGSAEWLGEGSSMSGV-VGTPYYVAPEVIMGREY 581
Query: 261 SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSK--EKFSPEFCSFVD 318
D+WS G+ L G FP E + +L PS S +
Sbjct: 582 DEKVDVWSSGVILYAMLAG-FPPFYGESAPEIFESVLRANLRFPSLIFSSVSAPAKDLLR 758
Query: 319 ACLQKDPDNRPTAEQLLLHPFI 340
+ +DP NR +A Q L HP+I
Sbjct: 759 KMISRDPSNRISAHQALRHPWI 824
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,072,857
Number of Sequences: 63676
Number of extensions: 338334
Number of successful extensions: 2748
Number of sequences better than 10.0: 715
Number of HSP's better than 10.0 without gapping: 2351
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2399
length of query: 519
length of database: 12,639,632
effective HSP length: 102
effective length of query: 417
effective length of database: 6,144,680
effective search space: 2562331560
effective search space used: 2562331560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146560.10


