

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.11 - phase: 0
(367 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
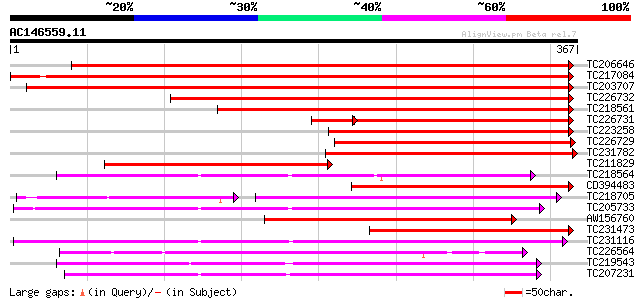
Score E
Sequences producing significant alignments: (bits) Value
TC206646 533 e-152
TC217084 510 e-145
TC203707 506 e-144
TC226732 371 e-103
TC218561 353 9e-98
TC226731 207 1e-64
TC223258 239 1e-63
TC226729 239 2e-63
TC231782 228 3e-60
TC211829 226 1e-59
TC218564 224 6e-59
CD394483 212 2e-55
TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g7... 132 5e-54
TC205733 204 5e-53
AW156760 197 5e-51
TC231473 196 2e-50
TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrola... 194 4e-50
TC226564 191 4e-49
TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase E... 191 4e-49
TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, pa... 190 1e-48
>TC206646
Length = 1178
Score = 533 bits (1373), Expect = e-152
Identities = 251/325 (77%), Positives = 290/325 (89%)
Frame = +1
Query: 41 DNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLV 100
D+GNN+FL TTARADAPPYGID+PTH PTGRFSNGLNIPDL S LGL+P+LPYLSPLLV
Sbjct: 7 DSGNNDFLVTTARADAPPYGIDYPTHRPTGRFSNGLNIPDLISLELGLKPTLPYLSPLLV 186
Query: 101 GEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNK 160
GEKLL+GANFASAG+GILNDTG QFL IIHI KQL LF++YQ++LS IGAEG + L N+
Sbjct: 187 GEKLLIGANFASAGIGILNDTGIQFLNIIHIQKQLKLFHEYQERLSLHIGAEGTRNLXNR 366
Query: 161 AIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTG 220
A+VLI LGGNDFVNNYYLVP+SARSRQFSLP+YV YLISEY+K+L+RLYDLGARRVLVTG
Sbjct: 367 ALVLITLGGNDFVNNYYLVPYSARSRQFSLPDYVRYLISEYRKVLRRLYDLGARRVLVTG 546
Query: 221 TGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMH 280
TGPMGC PAELA +SR GDCD EL RAASL+NPQLVQM+ LN+E+G DVFIA NA +MH
Sbjct: 547 TGPMGCVPAELATRSRTGDCDVELQRAASLFNPQLVQMLNGLNQELGADVFIAANAQRMH 726
Query: 281 MDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRII 340
MDF++NP+A+GFVT+K ACCGQG +NG+GLCTP S LCPNR+LYAFWD FHPSEKASRII
Sbjct: 727 MDFVSNPRAYGFVTSKIACCGQGPYNGVGLCTPTSNLCPNRDLYAFWDPFHPSEKASRII 906
Query: 341 VQQMFIGSNLYMNPMNLSTVLAMDS 365
VQQ+ G+ YM+PMNLST++A+DS
Sbjct: 907 VQQILRGTTEYMHPMNLSTIMAIDS 981
>TC217084
Length = 1310
Score = 510 bits (1314), Expect = e-145
Identities = 245/366 (66%), Positives = 300/366 (81%), Gaps = 1/366 (0%)
Frame = +3
Query: 1 MASSLMLC-CSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPY 59
+ SS LC C I +I++ V A+ RAFFVFGDS+ DNGNNNFL TTARAD+ PY
Sbjct: 81 LVSSKFLCLCLLITLISIIVA---PQAEAARAFFVFGDSLVDNGNNNFLATTARADSYPY 251
Query: 60 GIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILN 119
GID +H +GRFSNGLN+PDL SE++G EP+LPYLSP L GE+LLVGANFASAG+GILN
Sbjct: 252 GIDSASHRASGRFSNGLNMPDLISEKIGSEPTLPYLSPQLNGERLLVGANFASAGIGILN 431
Query: 120 DTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLV 179
DTG QF+ II I +QL F QYQQ++SA IG E + LVNKA+VLI LGGNDFVNNYYLV
Sbjct: 432 DTGIQFINIIRITEQLAYFKQYQQRVSALIGEEQTRNLVNKALVLITLGGNDFVNNYYLV 611
Query: 180 PFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGD 239
PFSARSR+++LP+YV +LISEY+KIL LY+LGARRVLVTGTGP+GC PAELA+ S+NG+
Sbjct: 612 PFSARSREYALPDYVVFLISEYRKILANLYELGARRVLVTGTGPLGCVPAELAMHSQNGE 791
Query: 240 CDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDAC 299
C EL RA +L+NPQLVQ++ +LN +IG DVFI+ NA MH+DF++NP+A+GFVT+K AC
Sbjct: 792 CATELQRAVNLFNPQLVQLLHELNTQIGSDVFISANAFTMHLDFVSNPQAYGFVTSKVAC 971
Query: 300 CGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLST 359
CGQG +NGIGLCTP S LCPNR+LYAFWD FHPSE+A+R+IV + GS YM+PMNLST
Sbjct: 972 CGQGAYNGIGLCTPASNLCPNRDLYAFWDPFHPSERANRLIVDKFMTGSTEYMHPMNLST 1151
Query: 360 VLAMDS 365
++A+DS
Sbjct: 1152IIALDS 1169
>TC203707
Length = 1394
Score = 506 bits (1303), Expect = e-144
Identities = 239/354 (67%), Positives = 296/354 (83%)
Frame = +1
Query: 12 ILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGR 71
+L+ + V L A+ +RAFFVFGDS+ DNGNNNFL TTARADAPPYGID+PT PTGR
Sbjct: 52 VLLGIVLVVVRLKGAEAQRAFFVFGDSLVDNGNNNFLATTARADAPPYGIDYPTGRPTGR 231
Query: 72 FSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHI 131
FSNG NIPD S+ LG E +LPYL P L GE+LLVGANFASAG+GILNDTG QF+ II I
Sbjct: 232 FSNGYNIPDFISQSLGAESTLPYLDPELDGERLLVGANFASAGIGILNDTGIQFVNIIRI 411
Query: 132 GKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLP 191
+QL+ + +YQQ++S IG E ++L+N A+VLI LGGNDFVNNYYLVP+SARSRQ++LP
Sbjct: 412 YRQLEYWEEYQQRVSGLIGPEQTERLINGALVLITLGGNDFVNNYYLVPYSARSRQYNLP 591
Query: 192 NYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLY 251
+YV Y+ISEYKK+L+RLY++GARRVLVTGTGP+GC PAELA +S NGDC AEL +AA+L+
Sbjct: 592 DYVKYIISEYKKVLRRLYEIGARRVLVTGTGPLGCVPAELAQRSTNGDCSAELQQAAALF 771
Query: 252 NPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLC 311
NPQLVQ+I QLN EIG +VF+ VN +MH+DFI+NP+ +GFVT+K ACCGQG +NG+GLC
Sbjct: 772 NPQLVQIIRQLNSEIGSNVFVGVNTQQMHIDFISNPQRYGFVTSKVACCGQGPYNGLGLC 951
Query: 312 TPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDS 365
TP S LCPNR+ YAFWD FHP+E+A+RIIVQQ+ G++ YM PMNLST++A+DS
Sbjct: 952 TPASNLCPNRDSYAFWDPFHPTERANRIIVQQILSGTSEYMYPMNLSTIMALDS 1113
>TC226732
Length = 985
Score = 371 bits (953), Expect = e-103
Identities = 177/261 (67%), Positives = 216/261 (81%)
Frame = +3
Query: 105 LVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVL 164
LVGANFASAG+GILNDTG QF+ +I + +QL+ F +YQ ++SA IGA AK LV +A+VL
Sbjct: 9 LVGANFASAGIGILNDTGVQFVNVIRMYRQLEYFKEYQNRVSAIIGASEAKNLVKQALVL 188
Query: 165 IMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPM 224
I +GGNDFVNNY+LVP SARS+Q+ LP YV YLISEY+K+LQRLYDLGARRVLVTGTGP+
Sbjct: 189 ITVGGNDFVNNYFLVPNSARSQQYPLPAYVKYLISEYQKLLQRLYDLGARRVLVTGTGPL 368
Query: 225 GCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFI 284
GC P+ELA + RNG C EL +AA+L+NPQL QM+ QLNR+I DVFIA N K H DF+
Sbjct: 369 GCVPSELAQRGRNGQCVPELQQAAALFNPQLEQMLLQLNRKIATDVFIAANTGKAHNDFV 548
Query: 285 TNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQM 344
TN + FGFVT++ ACCGQG +NGIGLCT +S LC NR+ YAFWDAFHPSEKA+R+IV+++
Sbjct: 549 TNAQQFGFVTSQVACCGQGPYNGIGLCTALSNLCSNRDQYAFWDAFHPSEKANRLIVEEI 728
Query: 345 FIGSNLYMNPMNLSTVLAMDS 365
GS YMNPMNLST+LA+DS
Sbjct: 729 MSGSKAYMNPMNLSTILALDS 791
>TC218561
Length = 1149
Score = 353 bits (905), Expect = 9e-98
Identities = 162/231 (70%), Positives = 200/231 (86%)
Frame = +2
Query: 135 LDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYV 194
L+ F +YQQ++SA +G E K+LVN A+VLI GGNDFVNNYYLVP SARSRQF+LP+YV
Sbjct: 5 LEYFQEYQQRVSALVGDEKTKELVNGALVLITCGGNDFVNNYYLVPNSARSRQFALPDYV 184
Query: 195 TYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQ 254
TY+ISEYKK+L+RLYDLGARRVLVTGTGP+GC PAELAL+ RNG+C EL RA++LYNPQ
Sbjct: 185 TYVISEYKKVLRRLYDLGARRVLVTGTGPLGCVPAELALRGRNGECSEELQRASALYNPQ 364
Query: 255 LVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPI 314
LV+MI QLN+E+G DVF+A N MH DF+TNP+A+GF+T+K ACCGQG FNG+GLCT +
Sbjct: 365 LVEMIKQLNKEVGSDVFVAANTQLMHDDFVTNPQAYGFITSKVACCGQGPFNGLGLCTVV 544
Query: 315 SKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDS 365
S LCPNR+ +AFWD FHPSEKA+R+IVQQ+ G++ YM+PMNLST+LA+DS
Sbjct: 545 SNLCPNRHEFAFWDPFHPSEKANRLIVQQIMSGTSKYMHPMNLSTILALDS 697
>TC226731
Length = 782
Score = 207 bits (526), Expect(2) = 1e-64
Identities = 92/142 (64%), Positives = 115/142 (80%)
Frame = +1
Query: 224 MGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDF 283
+GC P+ELA + RNG C EL +AA+L+NPQL QM+ QLNR+IG DVFIA N K H DF
Sbjct: 91 LGCVPSELAQRGRNGQCAPELQQAAALFNPQLEQMLLQLNRKIGSDVFIAANTGKAHNDF 270
Query: 284 ITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQ 343
+TNP+ FGFVT++ ACCGQG +NG+GLCT +S LC NR YAFWDAFHPSEKA+R+IV++
Sbjct: 271 VTNPRQFGFVTSQVACCGQGPYNGLGLCTALSNLCSNRETYAFWDAFHPSEKANRLIVEE 450
Query: 344 MFIGSNLYMNPMNLSTVLAMDS 365
+ GS YMNPMNLST+LA+D+
Sbjct: 451 IMSGSKAYMNPMNLSTILALDA 516
Score = 58.2 bits (139), Expect(2) = 1e-64
Identities = 26/30 (86%), Positives = 30/30 (99%)
Frame = +3
Query: 196 YLISEYKKILQRLYDLGARRVLVTGTGPMG 225
YLISEY+K+L+RLYDLGARRVLVTGTGP+G
Sbjct: 6 YLISEYQKLLKRLYDLGARRVLVTGTGPLG 95
>TC223258
Length = 672
Score = 239 bits (611), Expect = 1e-63
Identities = 106/159 (66%), Positives = 135/159 (84%)
Frame = +3
Query: 207 RLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREI 266
RLY+LGARRVLVTG+GP+GCAPAELA++ +NG+C A+L RAASLYNPQL QM+ +LN++I
Sbjct: 3 RLYNLGARRVLVTGSGPLGCAPAELAMRGKNGECSADLQRAASLYNPQLEQMLLELNKKI 182
Query: 267 GDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAF 326
G DVFIA N MH DFITNP A+GF T+K ACCGQG +NG+GLC P+S LCPNR+L+AF
Sbjct: 183 GSDVFIAANTALMHNDFITNPNAYGFNTSKVACCGQGPYNGMGLCLPVSNLCPNRDLHAF 362
Query: 327 WDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDS 365
WD FHP+EKA++++V+Q+ GS YM PMNLST+L +D+
Sbjct: 363 WDPFHPTEKANKLVVEQIMSGSTKYMKPMNLSTILTLDA 479
>TC226729
Length = 696
Score = 239 bits (609), Expect = 2e-63
Identities = 106/156 (67%), Positives = 133/156 (84%)
Frame = +2
Query: 211 LGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDV 270
LGARRVLVTGTGP+GC P+ELA + RNG C AEL +AA L+NPQL QM+ QLNR+IG D
Sbjct: 2 LGARRVLVTGTGPLGCVPSELAQRGRNGQCAAELQQAAELFNPQLEQMLLQLNRKIGKDT 181
Query: 271 FIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAF 330
FIA N KMH +F+TNP+ FGF+T++ ACCGQG +NG+GLCTP+S LCPNR+ YAFWDAF
Sbjct: 182 FIAANTGKMHNNFVTNPQQFGFITSQIACCGQGPYNGLGLCTPLSNLCPNRDQYAFWDAF 361
Query: 331 HPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSM 366
HPSEKA+R+IV+++ GS +YMNPMNLST+LA+D++
Sbjct: 362 HPSEKANRLIVEEIMSGSKIYMNPMNLSTILALDAI 469
>TC231782
Length = 701
Score = 228 bits (581), Expect = 3e-60
Identities = 101/163 (61%), Positives = 135/163 (81%)
Frame = +2
Query: 205 LQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNR 264
L RLY+LGARRVLVTGTGP+GC PA+LA +S NG+C EL +AA ++NP LVQM ++N
Sbjct: 2 LMRLYELGARRVLVTGTGPLGCVPAQLATRSSNGECVPELQQAAQIFNPLLVQMTREINS 181
Query: 265 EIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLY 324
++G DVF+AVNA +M+M+FIT+P+ FGFVT+K ACCGQGRFNG+GLCT +S LCPNR++Y
Sbjct: 182 QVGSDVFVAVNAFQMNMNFITDPQRFGFVTSKIACCGQGRFNGVGLCTSLSNLCPNRDIY 361
Query: 325 AFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLSTVLAMDSMV 367
AFWD +HPS++A IV+ +F G++ M PMNLST++A+DS +
Sbjct: 362 AFWDPYHPSQRALGFIVRDIFSGTSDIMTPMNLSTIMAIDSNI 490
>TC211829
Length = 446
Score = 226 bits (576), Expect = 1e-59
Identities = 110/148 (74%), Positives = 127/148 (85%)
Frame = +3
Query: 62 DFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDT 121
D+PTH TGRFSNGLNIPD+ SE++G EP+LPYLS L GE+LLVGANFASAG+GILNDT
Sbjct: 3 DYPTHRATGRFSNGLNIPDIISEKIGSEPTLPYLSRELDGERLLVGANFASAGIGILNDT 182
Query: 122 GFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPF 181
G QF+ II I +QL F QYQQ++SA IG E ++LVN+A+VLI LGGNDFVNNYYLVPF
Sbjct: 183 GIQFINIIRISRQLQYFEQYQQRVSALIGPEQTQRLVNQALVLITLGGNDFVNNYYLVPF 362
Query: 182 SARSRQFSLPNYVTYLISEYKKILQRLY 209
SARSRQF+LPNYV YLISEY+KIL RLY
Sbjct: 363 SARSRQFALPNYVVYLISEYRKILVRLY 446
>TC218564
Length = 1398
Score = 224 bits (570), Expect = 6e-59
Identities = 122/314 (38%), Positives = 179/314 (56%), Gaps = 4/314 (1%)
Frame = +1
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A VFGDS D+GNNN + T +++ PYG DF PTGRF NG PD +E G++
Sbjct: 91 AVIVFGDSSVDSGNNNVIATVLKSNFKPYGRDFEGDRPTGRFCNGRVPPDFIAEAFGIKR 270
Query: 91 SLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
++P YL P + G FASAG G N T L +I + K+++ + +YQ KL +
Sbjct: 271 TVPAYLDPAYTIQDFATGVCFASAGTGYDNATS-AVLNVIPLWKEIEYYKEYQAKLRTHL 447
Query: 150 GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLY 209
G E A +++++A+ L+ LG NDF+ NYY+ P R F++ Y +L+ + ++ LY
Sbjct: 448 GVEKANKIISEALYLMSLGTNDFLENYYVFP--TRRLHFTVSQYQDFLLRIAENFVRELY 621
Query: 210 DLGARRVLVTGTGPMGCAPAELALKSRNGD--CDAELMRAASLYNPQLVQMITQLNREIG 267
LG R++ +TG GP+GC P E A GD C+ E A +N +L +IT+LNRE+
Sbjct: 622 ALGVRKLSITGLGPVGCLPLERATNIL-GDHGCNQEYNDVALSFNRKLENVITKLNRELP 798
Query: 268 DDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKL-CPNRNLYAF 326
++ NA+ + D IT P +GF + ACC G F LC+ + L C + Y F
Sbjct: 799 RLKALSANAYSIVNDIITKPSTYGFEVVEKACCSTGTFEMSYLCSDKNPLTCTDAEKYVF 978
Query: 327 WDAFHPSEKASRII 340
WDAFHP+EK +RI+
Sbjct: 979 WDAFHPTEKTNRIV 1020
>CD394483
Length = 605
Score = 212 bits (539), Expect = 2e-55
Identities = 95/144 (65%), Positives = 116/144 (79%)
Frame = -1
Query: 222 GPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHM 281
GPM C PAELA++ NG C AEL RAASLYNPQL MI LN++IG DVFIA N MH
Sbjct: 605 GPMACVPAELAMRGTNGGCSAELQRAASLYNPQLTHMIQGLNKKIGKDVFIAANTALMHN 426
Query: 282 DFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIV 341
DF++NP A+GF T++ ACCGQG +NGIGLCTP+S LCPNRNL+AFWD FHPSEK++R+IV
Sbjct: 425 DFVSNPAAYGFTTSQIACCGQGPYNGIGLCTPLSDLCPNRNLHAFWDPFHPSEKSNRLIV 246
Query: 342 QQMFIGSNLYMNPMNLSTVLAMDS 365
+Q+ GS YM PMNLSTV+++D+
Sbjct: 245 EQIMSGSKRYMKPMNLSTVISLDA 174
>TC218705 weakly similar to GB|AAO63389.1|28950931|BT005325 At1g71250
{Arabidopsis thaliana;} , partial (78%)
Length = 1324
Score = 132 bits (331), Expect(2) = 5e-54
Identities = 65/199 (32%), Positives = 106/199 (52%), Gaps = 1/199 (0%)
Frame = +1
Query: 160 KAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVT 219
K+I +++ G ND++NNY L SR ++ ++ L++ Y + + L+ +G R+ +
Sbjct: 529 KSIAVVVTGSNDYINNYLLPGLYGSSRNYTAQDFGNLLVNSYVRQILALHSVGLRKFFLA 708
Query: 220 GTGPMGCAPA-ELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHK 278
G GP+GC P A + G C + + +N L M+ QLNR + +F+ N ++
Sbjct: 709 GIGPLGCIPTLRAAALAPTGRCVDLVNQMVGTFNEGLRSMVDQLNRNHPNAIFVYGNTYR 888
Query: 279 MHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASR 338
+ D + NP AF F ACCG GR G C P+ C +RN Y FWDAFHP+E A+
Sbjct: 889 VFGDILNNPAAFAFNVVDRACCGIGRNRGQLTCLPLQFPCTSRNQYVFWDAFHPTESATY 1068
Query: 339 IIVQQMFIGSNLYMNPMNL 357
+ ++ G+ P+N+
Sbjct: 1069VFAWRVVNGAPDDSYPINM 1125
Score = 97.4 bits (241), Expect(2) = 5e-54
Identities = 56/148 (37%), Positives = 84/148 (55%), Gaps = 4/148 (2%)
Frame = +2
Query: 5 LMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFP 64
L+LC SY + + +Q FVFGDS+ + GNNNFL T ARA+ PYGIDF
Sbjct: 83 LVLCSSY------GIAEVKSQSQKVSGLFVFGDSLVEVGNNNFLNTIARANYFPYGIDF- 241
Query: 65 THEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGEKLLVGANFASAGVGILNDTGFQ 124
+ TGRFSNG ++ D + LG+ P+ P VG ++L G N+ASA GIL+++G
Sbjct: 242 SKSSTGRFSNGKSLIDFIGDLLGIPSPTPFADPSTVGTRILYGVNYASASAGILDESGRH 421
Query: 125 FLQIIHIGKQL----DLFNQYQQKLSAQ 148
+ + +Q+ + NQY+ ++ +
Sbjct: 422 YGDRYSLSQQVLNFENTLNQYRTMMNGK 505
>TC205733
Length = 1377
Score = 204 bits (519), Expect = 5e-53
Identities = 123/347 (35%), Positives = 181/347 (51%), Gaps = 3/347 (0%)
Frame = +2
Query: 3 SSLMLCCSYILMINLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFLTTTARADAPPYGID 62
++ +L CS+I+++ L + + A VFGDS D GNNNF+ T AR++ PYG D
Sbjct: 77 TTTLLLCSHIVVL-LLLSLVAETSAKVSAVIVFGDSSVDAGNNNFIPTIARSNFQPYGRD 253
Query: 63 FPTHEPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILNDT 121
F + TGRF NG D SE GL+P +P YL P G FASA G N T
Sbjct: 254 FEGGKATGRFCNGRIPTDFISESFGLKPYVPAYLDPKYNISDFASGVTFASAATGYDNAT 433
Query: 122 GFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPF 181
L +I + KQL+ + YQ+ LSA +G AK + +A+ L+ LG NDF+ NYY +P
Sbjct: 434 S-DVLSVIPLWKQLEYYKGYQKNLSAYLGESKAKDTIAEALHLMSLGTNDFLENYYTMP- 607
Query: 182 SARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNG-DC 240
R+ QF+ Y +L + ++ LY LGAR+V + G PMGC P E G DC
Sbjct: 608 -GRASQFTPQQYQNFLAGIAENFIRSLYGLGARKVSLGGLPPMGCLPLERTTSIAGGNDC 784
Query: 241 DAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACC 300
A A +N +L + +LN+E+ + N + + + I P+ +GF + ACC
Sbjct: 785 VARYNNIALEFNNRLKNLTIKLNQELPGLKLVFSNPYYIMLSIIKRPQLYGFESTSVACC 964
Query: 301 GQGRFNGIGLCTPISKL-CPNRNLYAFWDAFHPSEKASRIIVQQMFI 346
G F C+ C + + Y FWD+FHP+E + I+ + + +
Sbjct: 965 ATGMFEMGYACSRGQMFSCTDASKYVFWDSFHPTEMTNSIVAKYVVL 1105
>AW156760
Length = 493
Score = 197 bits (502), Expect = 5e-51
Identities = 93/163 (57%), Positives = 125/163 (76%)
Frame = +2
Query: 166 MLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMG 225
+L G+D+VNNYY V +S SRQ++L +Y+ +LISEY+KIL LY+L A+ VLVT TGP+
Sbjct: 2 ILCGDDYVNNYY*VTWSGISRQYTLSDYMVFLISEYRKILANLYELRAQMVLVTCTGPLC 181
Query: 226 CAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFIT 285
C P+ELA+ S N +C EL R+ L+NPQLVQ++ +LN +IG DVFI+ A MH+DF++
Sbjct: 182 CVPSELAMHSHNRECATELQRSRDLFNPQLVQLLHELNTQIGSDVFISEYAFTMHLDFVS 361
Query: 286 NPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWD 328
NP+A+GFVT+K ACCGQ +NGI LCTP LCP+R+ YAF D
Sbjct: 362 NPQAYGFVTSKVACCGQRAYNGIRLCTPAVNLCPHRDGYAFSD 490
>TC231473
Length = 656
Score = 196 bits (497), Expect = 2e-50
Identities = 86/132 (65%), Positives = 112/132 (84%)
Frame = +2
Query: 234 KSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFV 293
+SRNG+C AEL +A++L+NPQLVQ++ QLN EIG DVFI+ NA + +MDFI+NP+A+GF+
Sbjct: 5 RSRNGECAAELQQASALFNPQLVQLVNQLNSEIGSDVFISANAFQSNMDFISNPQAYGFI 184
Query: 294 TAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMN 353
T+K ACCGQG +NGIGLCTP S LCPNR++YAFWD FHPSE+A+R+IV IG + YM+
Sbjct: 185 TSKVACCGQGPYNGIGLCTPASNLCPNRDVYAFWDPFHPSERANRLIVDTFMIGDSKYMH 364
Query: 354 PMNLSTVLAMDS 365
PMNLST+L +DS
Sbjct: 365 PMNLSTMLLLDS 400
>TC231116 similar to UP|Q9FFC6 (Q9FFC6) GDSL-motif lipase/hydrolase-like
protein, partial (94%)
Length = 1541
Score = 194 bits (494), Expect = 4e-50
Identities = 126/363 (34%), Positives = 178/363 (48%), Gaps = 4/363 (1%)
Frame = +1
Query: 3 SSLMLCCSYILMINLFVGFDLAYAQPK-RAFFVFGDSVADNGNNNFLTTTARADAPPYGI 61
SS+ S++ L V ++ QP A F FGDS+ D GNNN T +A+ PPYG
Sbjct: 358 SSMGYSRSFLASFLLAVLLNVTNGQPLVPAIFTFGDSIVDVGNNNHQLTIVKANFPPYGR 537
Query: 62 DFPTHEPTGRFSNGLNIPDLTSERLGLEPSLP-YLSPLLVGEKLLVGANFASAGVGILND 120
DF H PTGRF NG D ++ LG P YL+ G+ LL GANFASA G
Sbjct: 538 DFENHFPTGRFCNGKLATDFIADILGFTSYQPAYLNLKTKGKNLLNGANFASASSGYFEL 717
Query: 121 TGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVP 180
T + I + KQL+ + + Q KL G A +++ AI LI G +DFV NYY+ P
Sbjct: 718 TS-KLYSSIPLSKQLEYYKECQTKLVEAAGQSSASSIISDAIYLISAGTSDFVQNYYINP 894
Query: 181 FSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRN-GD 239
++ ++ + L+ Y +Q LY LGARR+ VT P+GC PA + L + +
Sbjct: 895 L--LNKLYTTDQFSDTLLRCYSNFIQSLYALGARRIGVTSLPPIGCLPAVITLFGAHINE 1068
Query: 240 CDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDAC 299
C L A +N +L L + + + ++ D T P GF A+ AC
Sbjct: 1069CVTSLNSDAINFNEKLNTTSQNLKNMLPGLNLVVFDIYQPLYDLATKPSENGFFEARKAC 1248
Query: 300 CGQGRFNGIGLCTPIS-KLCPNRNLYAFWDAFHPSEKASRIIVQQMFIGSNLYMNPMNLS 358
CG G LC S C N + Y FWD FHPSE A++++ ++ ++ N S
Sbjct: 1249CGTGLIEVSILCNKKSIGTCANASEYVFWDGFHPSEAANKVLADELITSGISLIS*SNTS 1428
Query: 359 TVL 361
V+
Sbjct: 1429RVI 1437
>TC226564
Length = 1337
Score = 191 bits (485), Expect = 4e-49
Identities = 108/306 (35%), Positives = 169/306 (54%), Gaps = 3/306 (0%)
Frame = +3
Query: 33 FVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSL 92
F+FGDS++D+GNNN L TT++++ PYGIDFP PTGR++NG D+ ++ LG E +
Sbjct: 147 FIFGDSMSDSGNNNELPTTSKSNFRPYGIDFPLG-PTGRYTNGRTEIDIITQFLGFEKFI 323
Query: 93 PYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAE 152
P + G +L G N+AS G GI N+TG+ + I +G QL +++ ++G+
Sbjct: 324 PPFANTS-GSDILKGVNYASGGSGIRNETGWHYGAAIGLGLQLANHRVIVSEIATKLGSP 500
Query: 153 G-AKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDL 211
A+Q + K + + +G ND++ NY+L PF S +++ + LI E LQ L+D+
Sbjct: 501 DLARQYLEKCLYYVNIGSNDYMGNYFLPPFYPTSTIYTIEEFTQVLIEELSLNLQALHDI 680
Query: 212 GARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREI--GDD 269
GAR+ + G G +GC P ++ NG C E AA +N +L + Q N + +
Sbjct: 681 GARKYALAGLGLIGCTPGMVSAHGTNGSCAEEQNLAAFNFNNKLKARVDQFNNDFYYANS 860
Query: 270 VFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDA 329
FI +N + ++ +GF + CC G G C P + C NRN Y F+DA
Sbjct: 861 KFIFINTQALAIEL---RDKYGFPVPETPCCLPGL---TGECVPDQEPCYNRNDYVFFDA 1022
Query: 330 FHPSEK 335
FHP+E+
Sbjct: 1023FHPTEQ 1040
>TC219543 weakly similar to UP|Q94CH8 (Q94CH8) Family II lipase EXL1, partial
(49%)
Length = 1085
Score = 191 bits (485), Expect = 4e-49
Identities = 107/318 (33%), Positives = 167/318 (51%), Gaps = 4/318 (1%)
Frame = +2
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDF-PTHEPTGRFSNGLNIPDLTSERLGLE 89
A VFGDS+ D GNNN++TT A+ + PYG DF ++PTGRFSNGL D+ + + G++
Sbjct: 122 AVIVFGDSIVDTGNNNYITTIAKCNFLPYGRDFGGGNQPTGRFSNGLTPSDIIAAKFGVK 301
Query: 90 PSL-PYLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQ 148
L PYL P L + LL G +FAS G + + + + QLD F +Y+ K+
Sbjct: 302 ELLPPYLDPKLQPQDLLTGVSFASGASG-YDPLTSKIASALSLSDQLDTFREYKNKIMEI 478
Query: 149 IGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRL 208
+G +++K+I ++ G ND N Y++ R ++ + Y + S+ LQ L
Sbjct: 479 VGENRTATIISKSIYILCTGSNDITNTYFV-----RGGEYDIQAYTDLMASQATNFLQEL 643
Query: 209 YDLGARRVLVTGTGPMGCAPAELALKSR-NGDCDAELMRAASLYNPQLVQMITQLNREIG 267
Y LGARR+ V G +GC P++ L C AA L+N +L + L ++
Sbjct: 644 YGLGARRIGVVGLPVLGCVPSQRTLHGGIFRACSDFENEAAVLFNSKLSSQMDALKKQFQ 823
Query: 268 DDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKL-CPNRNLYAF 326
+ F+ ++ + ++ I NP +GF CCG G+ LC + L C N + Y F
Sbjct: 824 EARFVYLDLYNPVLNLIQNPAKYGFEVMDQGCCGTGKLEVGPLCNHFTLLICSNTSNYIF 1003
Query: 327 WDAFHPSEKASRIIVQQM 344
WD+FHP+E A ++ Q+
Sbjct: 1004WDSFHPTEAAYNVVCTQV 1057
>TC207231 similar to UP|Q94CH6 (Q94CH6) Family II lipase EXL3, partial (70%)
Length = 1216
Score = 190 bits (482), Expect = 1e-48
Identities = 104/311 (33%), Positives = 159/311 (50%), Gaps = 2/311 (0%)
Frame = +3
Query: 36 GDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLP-Y 94
GDS+ D GNNN + T + D PPYG DF PTGRF NG DL E LG++ LP Y
Sbjct: 3 GDSIVDPGNNNKVKTLVKCDFPPYGKDFEGGIPTGRFCNGKIPSDLLVEELGIKELLPAY 182
Query: 95 LSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGA 154
L P L L+ G FAS G T + +I + +QLD+F +Y KL +G +
Sbjct: 183 LDPNLKPSDLVTGVCFASGASGYDPLTP-KIASVISMSEQLDMFKEYIGKLKHIVGEDRT 359
Query: 155 KQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGAR 214
K ++ + L++ G +D N Y++ R Q+ +P Y ++ ++ LY LGAR
Sbjct: 360 KFILANSFFLVVAGSDDIANTYFIA--RVRQLQYDIPAYTDLMLHSASNFVKELYGLGAR 533
Query: 215 RVLVTGTGPMGCAPAELALKSR-NGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIA 273
R+ V P+GC P++ L +C E AA L+N +L + + L + + +
Sbjct: 534 RIGVLSAPPIGCVPSQRTLAGGFQRECAEEYNYAAKLFNSKLSRELDALKHNLPNSRIVY 713
Query: 274 VNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNRNLYAFWDAFHPS 333
++ + MD I N + G+ CCG G+ LC P+ CP+ + Y FWD++HP+
Sbjct: 714 IDVYNPLMDIIVNYQRHGYKVVDRGCCGTGKLEVAVLCNPLGATCPDASQYVFWDSYHPT 893
Query: 334 EKASRIIVQQM 344
E R ++ Q+
Sbjct: 894 EGVYRQLIVQV 926
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,594,896
Number of Sequences: 63676
Number of extensions: 193717
Number of successful extensions: 1170
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 1049
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1052
length of query: 367
length of database: 12,639,632
effective HSP length: 99
effective length of query: 268
effective length of database: 6,335,708
effective search space: 1697969744
effective search space used: 1697969744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146559.11


