

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
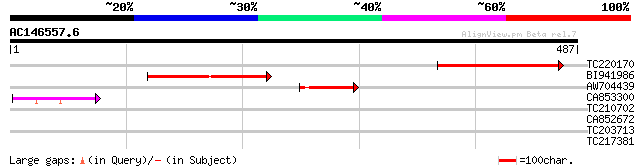
Query= AC146557.6 - phase: 0 /pseudo
(487 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220170 weakly similar to UP|SYD_SYNEL (Q8DJS8) Aspartyl-tRNA s... 150 9e-37
BI941986 146 2e-35
AW704439 59 5e-09
CA853300 54 1e-07
TC210702 similar to UP|Q9LJE2 (Q9LJE2) Lysyl-tRNA synthetase, pa... 30 3.2
CA852672 29 4.1
TC203713 similar to GB|AAP21358.1|30102880|BT006550 At5g19590 {A... 29 5.4
TC217381 similar to UP|O82469 (O82469) Protein phosphatase-2C , ... 28 7.1
>TC220170 weakly similar to UP|SYD_SYNEL (Q8DJS8) Aspartyl-tRNA synthetase
(Aspartate--tRNA ligase) (AspRS) , partial (32%)
Length = 939
Score = 150 bits (380), Expect = 9e-37
Identities = 78/109 (71%), Positives = 86/109 (78%), Gaps = 1/109 (0%)
Frame = +3
Query: 368 DIYNEAEKYGAKGLTTLWITKNG-IGGFSSLVSSMDSATIEELLRRCSAGPSDLILFHAG 426
DIYNEA K GAKGL L + +G I G S+LVSSMD T LLRRCS+GPSDLILF G
Sbjct: 39 DIYNEALKSGAKGLPFLKVLDDGNIEGISALVSSMDPTTRGNLLRRCSSGPSDLILFAVG 218
Query: 427 HHASVNKTLGHLRVYVAHKLGLIDHAKHSILWITDFPMFKWDDSKQRLE 475
HHASVNKTL LRVYVAH LGLID +HSILWITDFPMF+W+D +QRLE
Sbjct: 219 HHASVNKTLDRLRVYVAHDLGLIDLGRHSILWITDFPMFEWNDPEQRLE 365
>BI941986
Length = 419
Score = 146 bits (368), Expect = 2e-35
Identities = 77/107 (71%), Positives = 89/107 (82%)
Frame = +1
Query: 119 NLRREYVAAIQGVVRSRSTQSINYEMKTGFIEVAANEVQELNSRNAKLTLIPELNSDDAE 178
+LR EYV AI+GVVRSR +SIN +M+TGFIEVAAN+VQ LNS N+KL + +DDA+
Sbjct: 7 DLRLEYVVAIEGVVRSRPDESINKKMQTGFIEVAANKVQLLNSVNSKLPFLVT-TADDAK 183
Query: 179 GSPNEEIRLRYRCLDLRKQQMNSNILLRHNVVKLIRRYLEDIHGFVE 225
S EEIRLRYRCLDLR+QQMN NILLRH VVKLIRRYLED+HGFVE
Sbjct: 184 DSLKEEIRLRYRCLDLRRQQMNFNILLRHKVVKLIRRYLEDVHGFVE 324
>AW704439
Length = 238
Score = 58.9 bits (141), Expect = 5e-09
Identities = 33/50 (66%), Positives = 38/50 (76%)
Frame = +2
Query: 250 QDLRAVHDRQPEFTQLDMEMAFTPLEDILSLNEELIRKVFLEIKGVELPN 299
++LRA DRQ EFTQLDMEM FTP ED+L+LNEELIRKV L +L N
Sbjct: 74 ENLRA--DRQLEFTQLDMEMTFTPYEDMLTLNEELIRKVRLHYL*SDLSN 217
>CA853300
Length = 479
Score = 54.3 bits (129), Expect = 1e-07
Identities = 39/97 (40%), Positives = 48/97 (49%), Gaps = 21/97 (21%)
Frame = +3
Query: 3 SLFLKALSLISIRRSPSLR-----KTKIPLPYYEKLKPKTKKAIT--------------- 42
SLFLKAL L++ R PSL IPL + A T
Sbjct: 186 SLFLKALPLMAFRTRPSLVFLRRVSNTIPLSLSRTRTVYSASASTQPPPSACASETLTSS 365
Query: 43 FKKPKPLNQSLQWISRTHHCGELSSNDVGKTV-RLCG 78
K P PL++ L+W++RT CGELSSNDVGK V + CG
Sbjct: 366 AKPPLPLSEPLEWVNRTAFCGELSSNDVGKIVFK*CG 476
>TC210702 similar to UP|Q9LJE2 (Q9LJE2) Lysyl-tRNA synthetase, partial (20%)
Length = 623
Score = 29.6 bits (65), Expect = 3.2
Identities = 29/81 (35%), Positives = 37/81 (44%), Gaps = 2/81 (2%)
Frame = +3
Query: 25 IPLPYYEKLKPK--TKKAITFKKPKPLNQSLQWISRTHHCGELSSNDVGKTVRLCGWAEL 82
I L E+L+ K A + K NQ LQ I R GE N V + G
Sbjct: 282 IRLKKVEELRSKGLDPYAYEWNKTHGANQ-LQDIYRDLGNGE-EKNSENDHVSVAGRIAA 455
Query: 83 HRSLGGVAFLILRDQTGIIQV 103
R+ G +AFL LRD +G IQ+
Sbjct: 456 RRAFGKLAFLTLRDDSGTIQL 518
>CA852672
Length = 672
Score = 29.3 bits (64), Expect = 4.1
Identities = 19/72 (26%), Positives = 37/72 (51%), Gaps = 5/72 (6%)
Frame = +3
Query: 370 YNEAEKYGAKGLTTLWITKNGIGGFSSLVSSMDSATIEELLRRCSAGPSDLIL-----FH 424
+N A Y K LT+ W+T+ FSSL + ++ A I+ +++ +A + +++ +
Sbjct: 225 WNAAGTYEEKNLTS-WVTEYLTARFSSLSTIIEDAQIDGSVKQAAADVASILIQTTAASN 401
Query: 425 AGHHASVNKTLG 436
G HA + G
Sbjct: 402 VGGHAQLTLARG 437
>TC203713 similar to GB|AAP21358.1|30102880|BT006550 At5g19590 {Arabidopsis
thaliana;} , partial (64%)
Length = 645
Score = 28.9 bits (63), Expect = 5.4
Identities = 14/33 (42%), Positives = 21/33 (63%)
Frame = +1
Query: 203 ILLRHNVVKLIRRYLEDIHGFVEPCHKALRYIS 235
+LL H++ IRR D+H V P +KA+ Y+S
Sbjct: 430 VLLLHHIYA-IRRIYFDVHTLVGPVNKAILYVS 525
>TC217381 similar to UP|O82469 (O82469) Protein phosphatase-2C , partial
(35%)
Length = 742
Score = 28.5 bits (62), Expect = 7.1
Identities = 39/164 (23%), Positives = 71/164 (42%), Gaps = 1/164 (0%)
Frame = +2
Query: 12 ISIRRSPSLRKTKIPLPYYEKLKPKTKKAITFKKPKPLNQSLQWISRTHHCGELSSNDVG 71
I I +P+ + KIP + + KTK +++ K Q S ++ G S
Sbjct: 2 IIIITNPNRNREKIPQKSQHRRRRKTKHSLSIKS---FWNIAQQFSALYNVGFGS----- 157
Query: 72 KTVRLCGWAELHRSLGGVAFLILRDQTGIIQVKTRLDEFPVAHSANKNLRREYVAAIQGV 131
V + G E HR++ A ++ + G++ VK FP S + +
Sbjct: 158 --VIV*GILE-HRNMVAEAEVVCQQNVGVLDVK----HFPNKVSDVHEIGDADANSPPSF 316
Query: 132 VRSRSTQSINYEMKTGFIEV-AANEVQELNSRNAKLTLIPELNS 174
R R+T+S++ E+ T +V +A+ + + +A L IP + S
Sbjct: 317 DRVRATESVSAELLTSQEDVKSADRISDAALESAVLQFIPCIRS 448
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,077,998
Number of Sequences: 63676
Number of extensions: 287736
Number of successful extensions: 1771
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1751
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1768
length of query: 487
length of database: 12,639,632
effective HSP length: 101
effective length of query: 386
effective length of database: 6,208,356
effective search space: 2396425416
effective search space used: 2396425416
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146557.6


