

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.5 - phase: 0
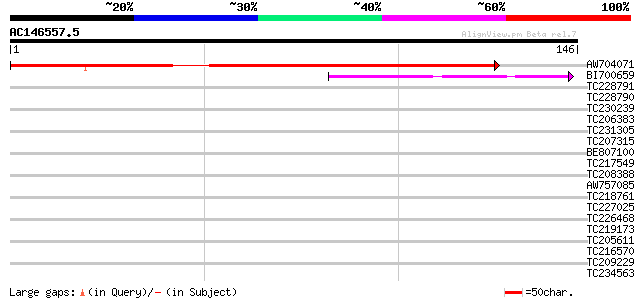
(146 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW704071 similar to GP|11558192|emb transcription factor (E2F) {... 170 3e-43
BI700659 similar to GP|8977833|emb| transcription factor E2F {Da... 52 1e-07
TC228791 34 0.022
TC228790 32 0.11
TC230239 weakly similar to UP|Q93Z73 (Q93Z73) At2g46080/T3F17.27... 32 0.11
TC206383 homologue to UP|Q9SDX4 (Q9SDX4) Dynamin homolog, partia... 30 0.32
TC231305 similar to UP|Q8LDC8 (Q8LDC8) Ethylene response factor ... 28 1.6
TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kina... 28 2.1
BE807100 weakly similar to SP|O60879|DIA2 Diaphanous protein hom... 27 2.7
TC217549 27 2.7
TC208388 similar to UP|Q8LNZ8 (Q8LNZ8) Abscisic acid insensitive... 27 2.7
AW757085 27 3.5
TC218761 similar to UP|Q9XIS4 (Q9XIS4) Starch branching enzyme ... 27 3.5
TC227025 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 27 3.5
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 27 3.5
TC219173 weakly similar to UP|NRG2_HUMAN (O14511) Pro-neuregulin... 27 3.5
TC205611 27 3.5
TC216570 similar to GB|AAO63420.1|28950993|BT005356 At5g28040 {A... 27 4.6
TC209229 similar to GB|AAS76730.1|45773852|BT012243 At5g21960 {A... 27 4.6
TC234563 similar to UP|Q6YZC6 (Q6YZC6) Cation exchanger-like pro... 27 4.6
>AW704071 similar to GP|11558192|emb transcription factor (E2F) {Chenopodium
rubrum}, partial (8%)
Length = 381
Score = 170 bits (430), Expect = 3e-43
Identities = 92/128 (71%), Positives = 96/128 (74%), Gaps = 2/128 (1%)
Frame = +2
Query: 1 MSGTRTPPATAAPDQIMQ--QRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVK 58
MS +R PPA PDQIM +RQLPFSSMKPPF+AA DYHR EAIVVK
Sbjct: 23 MSASRAPPAANPPDQIMHSMKRQLPFSSMKPPFVAAGDYHRHDA---------AEAIVVK 175
Query: 59 TPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQT 118
TP LKRKSE ADFEADSGDRMT G T AANS QTPVSGK GKGGKSSR+TK NR GTQT
Sbjct: 176 TPLLKRKSEVADFEADSGDRMTAGFTEAANSPFQTPVSGKTGKGGKSSRLTKGNRVGTQT 355
Query: 119 PGSNIGSP 126
PGSNIGSP
Sbjct: 356 PGSNIGSP 379
>BI700659 similar to GP|8977833|emb| transcription factor E2F {Daucus
carota}, partial (19%)
Length = 414
Score = 52.0 bits (123), Expect = 1e-07
Identities = 32/63 (50%), Positives = 36/63 (56%)
Frame = +1
Query: 83 STAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGPCRYDSS 142
S NS +TPVS K G+ K+ RS TP SN GSPS LTPA CRYDSS
Sbjct: 70 SNVTNNSPFKTPVSAKGGRTQKAKASK--GRSCPPTPISNAGSPSP--LTPASSCRYDSS 237
Query: 143 LGI 145
LG+
Sbjct: 238 LGL 246
>TC228791
Length = 584
Score = 34.3 bits (77), Expect = 0.022
Identities = 26/66 (39%), Positives = 32/66 (48%), Gaps = 3/66 (4%)
Frame = -2
Query: 77 DRMTPGSTAAANSSVQTPVS---GKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTP 133
DR PGS AAA ++P G G GG+ R + GT+TP S SP + P
Sbjct: 421 DRADPGSCAAAWPPRRSPTMRRRGTRG*GGRGCRSPAVSGGGTRTPRS---SP----VCP 263
Query: 134 AGPCRY 139
GP RY
Sbjct: 262 RGPGRY 245
>TC228790
Length = 1215
Score = 32.0 bits (71), Expect = 0.11
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 3/59 (5%)
Frame = -2
Query: 73 ADSGDRMTPGSTAAANSSVQTPV---SGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSG 128
A + D PGS AAA+ Q+P + G GG+ R + GT TP S+ P G
Sbjct: 350 AAARDLADPGSCAAASPPRQSPTRRRRERRG*GGRGCRSPAASGGGTHTPRSSPVCPRG 174
>TC230239 weakly similar to UP|Q93Z73 (Q93Z73) At2g46080/T3F17.27, partial
(84%)
Length = 1094
Score = 32.0 bits (71), Expect = 0.11
Identities = 18/34 (52%), Positives = 20/34 (57%)
Frame = +3
Query: 25 SSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVK 58
SS K L AD H +APD RR QDL E I V+
Sbjct: 546 SSKKLSDLNVADIHSWAPDFRRLQDLVNEEIRVR 647
>TC206383 homologue to UP|Q9SDX4 (Q9SDX4) Dynamin homolog, partial (46%)
Length = 1670
Score = 30.4 bits (67), Expect = 0.32
Identities = 21/62 (33%), Positives = 27/62 (42%), Gaps = 1/62 (1%)
Frame = +2
Query: 76 GDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGS-PSGNNLTPA 134
GD AAAN V S ++G G S + ++G GSN S + N L PA
Sbjct: 1076 GDDWRSAFDAAANGPVSRSGSSRSGSNGHSRHSSDPAQNGDVNSGSNSSSRRTPNRLPPA 1255
Query: 135 GP 136
P
Sbjct: 1256 PP 1261
>TC231305 similar to UP|Q8LDC8 (Q8LDC8) Ethylene response factor ERF1,
partial (45%)
Length = 856
Score = 28.1 bits (61), Expect = 1.6
Identities = 18/56 (32%), Positives = 27/56 (48%)
Frame = +3
Query: 23 PFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQLKRKSEAADFEADSGDR 78
P +P A+ +H F P R+ D + A + QL R++EAA E GD+
Sbjct: 60 PLQRKRPRRHASLWHHHFMP--RKKSDHQPRASE*EEEQLPRRAEAAVGEVRGGDK 221
>TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1063
Score = 27.7 bits (60), Expect = 2.1
Identities = 12/26 (46%), Positives = 13/26 (49%)
Frame = +1
Query: 103 GKSSRMTKCNRSGTQTPGSNIGSPSG 128
G SSR T C+ GT G PSG
Sbjct: 424 GTSSRKTSCSMKGTSLSSQTSGPPSG 501
>BE807100 weakly similar to SP|O60879|DIA2 Diaphanous protein homolog 2
(Diaphanous-related formin 2) (DRF2). [Human] {Homo
sapiens}, partial (3%)
Length = 385
Score = 27.3 bits (59), Expect = 2.7
Identities = 15/43 (34%), Positives = 19/43 (43%)
Frame = -3
Query: 93 TPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAG 135
TP A +G K R K + G + P G+P GN P G
Sbjct: 302 TPAGKPAARGQK--RRGKTQKKGGKGPTKRTGTPGGNPPPPGG 180
>TC217549
Length = 516
Score = 27.3 bits (59), Expect = 2.7
Identities = 19/47 (40%), Positives = 26/47 (54%), Gaps = 5/47 (10%)
Frame = +1
Query: 65 KSEAADFEADSGDRMTPGSTAAANSSVQTPV-----SGKAGKGGKSS 106
K EAA +D + GS A+ +++ + PV S K GKGGKSS
Sbjct: 130 KKEAAPKASDGASSKSKGS-ASKSATKKVPVQKPQESKKKGKGGKSS 267
>TC208388 similar to UP|Q8LNZ8 (Q8LNZ8) Abscisic acid insensitive 3-like
factor, partial (42%)
Length = 1236
Score = 27.3 bits (59), Expect = 2.7
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = +3
Query: 94 PVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPS 127
P + KAGK K+ T N S T +N G+PS
Sbjct: 894 PETKKAGKSQKNQHGTGTNASSTAGTAANNGTPS 995
>AW757085
Length = 431
Score = 26.9 bits (58), Expect = 3.5
Identities = 22/76 (28%), Positives = 33/76 (42%), Gaps = 12/76 (15%)
Frame = +2
Query: 66 SEAADFEADSGDRMTPGSTAAANSSVQTPVSGKAGKG---------GKSSRMTKCNRSG- 115
S AAD A D ++ GST+ A+ S K +G +SR+ + G
Sbjct: 2 SLAADLTASDTDSVSSGSTSGAHDSSGAAKGTKEPRGIVVSARFWQETNSRLRRLQDPGS 181
Query: 116 --TQTPGSNIGSPSGN 129
+ +P S IG P+ N
Sbjct: 182 PLSTSPASRIGVPNRN 229
>TC218761 similar to UP|Q9XIS4 (Q9XIS4) Starch branching enzyme , partial
(12%)
Length = 780
Score = 26.9 bits (58), Expect = 3.5
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 3/85 (3%)
Frame = -1
Query: 46 RNQDLETEAIVVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQT-PVSGKAGKGGK 104
R L TEA + + L + + E +S + + AAAN S T + KG
Sbjct: 528 RTTSLSTEAATIVSSNLSKLIPSVSTETNSPEIFVTSAFAAANGSFTTSKLLSSLSKGAD 349
Query: 105 SSRM--TKCNRSGTQTPGSNIGSPS 127
SS + T + + TP + I S S
Sbjct: 348 SSGIFATSAAANVSFTPSNLISSLS 274
>TC227025 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate
dehydrogenase, complete
Length = 2097
Score = 26.9 bits (58), Expect = 3.5
Identities = 15/51 (29%), Positives = 24/51 (46%)
Frame = +2
Query: 86 AANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNNLTPAGP 136
+A S TP S + S++ + + TP S PS + +TP+GP
Sbjct: 380 SAASPSSTPTSPPPSRRPSSAKRSPAASPSSPTPPSPHLPPSWSTMTPSGP 532
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 26.9 bits (58), Expect = 3.5
Identities = 20/67 (29%), Positives = 29/67 (42%), Gaps = 9/67 (13%)
Frame = -1
Query: 79 MTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCN---------RSGTQTPGSNIGSPSGN 129
+T STA+ NSS T G + S R T + ++ TQT ++ S S N
Sbjct: 540 ITCSSTASRNSSTSTSDRGTSASASASDRGTGSSACTP*A*KAQAQTQTQKASSTSSSSN 361
Query: 130 NLTPAGP 136
N + P
Sbjct: 360 NYSQKSP 340
>TC219173 weakly similar to UP|NRG2_HUMAN (O14511) Pro-neuregulin-2 precursor
(Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural-and
thymus-derived activator for ERBB kinases) (NTAK)
(Divergent of neuregulin 1) (DON-1)], partial (3%)
Length = 1120
Score = 26.9 bits (58), Expect = 3.5
Identities = 17/59 (28%), Positives = 24/59 (39%), Gaps = 5/59 (8%)
Frame = +2
Query: 10 TAAPDQIMQQRQLPFSSMKPP-----FLAAADYHRFAPDHRRNQDLETEAIVVKTPQLK 63
T+ P+ + Q LPF S+ PP F AA H P + + + P LK
Sbjct: 95 TSLPNSLPPQSLLPFQSLTPPLPLPEFSAAPPIHAPTPSKTKTTRRRSARRLPTNPSLK 271
>TC205611
Length = 672
Score = 26.9 bits (58), Expect = 3.5
Identities = 21/68 (30%), Positives = 32/68 (46%), Gaps = 5/68 (7%)
Frame = +3
Query: 56 VVKTPQLKRKSEAADFEADSGDRMTPGSTAAANSSVQTPVSGKA----GKGGKSSRMT-K 110
+V ++ KS+ D +S D T +A S +TP SGK+ K GK + T K
Sbjct: 69 IVTAKKIGNKSKNTDNSIESKDDDTSTPKPSAKSKQETPKSGKSKQETPKSGKFKQETPK 248
Query: 111 CNRSGTQT 118
+S +T
Sbjct: 249 SGKSKQET 272
>TC216570 similar to GB|AAO63420.1|28950993|BT005356 At5g28040 {Arabidopsis
thaliana;} , partial (26%)
Length = 1084
Score = 26.6 bits (57), Expect = 4.6
Identities = 17/68 (25%), Positives = 28/68 (41%), Gaps = 10/68 (14%)
Frame = +3
Query: 40 FAPDHRRNQDLETEAIVVKTPQLKRKSEAADFEADSG----------DRMTPGSTAAANS 89
F+P +N+ + + KTP+ R A E + G TP +TAAA +
Sbjct: 210 FSPGILKNETIFRNSTEKKTPKRSRPRSAVKIEPNDGSASNRDHNCISNTTPTATAAATN 389
Query: 90 SVQTPVSG 97
+ +G
Sbjct: 390 TTAAAAAG 413
>TC209229 similar to GB|AAS76730.1|45773852|BT012243 At5g21960 {Arabidopsis
thaliana;} , partial (38%)
Length = 1079
Score = 26.6 bits (57), Expect = 4.6
Identities = 16/56 (28%), Positives = 27/56 (47%)
Frame = -3
Query: 75 SGDRMTPGSTAAANSSVQTPVSGKAGKGGKSSRMTKCNRSGTQTPGSNIGSPSGNN 130
S ++ PGS+ S + KSS++ K +SGT++P + SGN+
Sbjct: 747 SSPKLKPGSSCKKKSRL------------KSSKLGKSEKSGTKSPDPKSENISGND 616
>TC234563 similar to UP|Q6YZC6 (Q6YZC6) Cation exchanger-like protein,
partial (50%)
Length = 798
Score = 26.6 bits (57), Expect = 4.6
Identities = 18/59 (30%), Positives = 27/59 (45%)
Frame = +2
Query: 8 PATAAPDQIMQQRQLPFSSMKPPFLAAADYHRFAPDHRRNQDLETEAIVVKTPQLKRKS 66
PA++ D+ + + +S P A H HR+ QD T+ +VV QLK S
Sbjct: 173 PASSTEDRAHEPAETGLTS---PVKLDATAHAHIEKHRKLQDDLTDEMVVLAKQLKESS 340
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.125 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,513,923
Number of Sequences: 63676
Number of extensions: 64992
Number of successful extensions: 395
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 388
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 392
length of query: 146
length of database: 12,639,632
effective HSP length: 88
effective length of query: 58
effective length of database: 7,036,144
effective search space: 408096352
effective search space used: 408096352
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146557.5


