

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146555.15 - phase: 0 /pseudo
(552 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
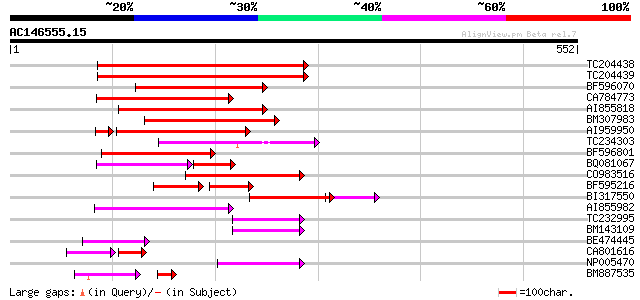
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 157 9e-39
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 157 9e-39
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 146 3e-35
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 142 4e-34
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 127 1e-29
BM307983 110 2e-24
AI959950 97 2e-20
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 95 9e-20
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 94 2e-19
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 67 2e-19
CO983516 91 2e-18
BF595216 56 4e-18
BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Ara... 83 5e-18
AI855982 82 6e-16
TC232995 59 6e-09
BM143109 57 3e-08
BE474445 55 6e-08
CA801616 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberos... 43 1e-07
NP005470 reverse transcriptase 54 2e-07
BM887535 similar to GP|14719283|gb Fourf gag/pol protein {Zea ma... 42 6e-07
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 157 bits (398), Expect = 9e-39
Identities = 84/207 (40%), Positives = 127/207 (60%), Gaps = 1/207 (0%)
Frame = +1
Query: 86 VSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVD 145
VS EPK+ EA+ + W MQ EL + W++V P + IG W+ K K N +
Sbjct: 3190 VSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEE 3369
Query: 146 GSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFL 205
G I R K LVA+GY QIEG+D+ +TF+ VA++ +IRL++ +A I L+Q+DV +AFL
Sbjct: 3370 GVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFL 3549
Query: 206 HGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATS 264
+G L+E+ Y+ P G V + P+ V +L K+LYGLK A R WYE+LT GY++
Sbjct: 3550 NGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGI 3729
Query: 265 NASLFTKKNLDSFIMLLVYVYDITLAG 291
+ +LF K++ ++ ++ +YV DI G
Sbjct: 3730 DKTLFVKQDAENLMIAQIYVDDIVFGG 3810
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 157 bits (398), Expect = 9e-39
Identities = 84/207 (40%), Positives = 127/207 (60%), Gaps = 1/207 (0%)
Frame = +1
Query: 86 VSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVD 145
VS EPK+ EA+ + W MQ EL + W++V P + IG W+ K K N +
Sbjct: 3187 VSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEE 3366
Query: 146 GSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFL 205
G I R K LVA+GY QIEG+D+ +TF+ VA++ +IRL++ +A I L+Q+DV +AFL
Sbjct: 3367 GVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFL 3546
Query: 206 HGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATS 264
+G L+E+VY+ P G + P+ V +L K+LYGLK A R WYE+LT GY++
Sbjct: 3547 NGYLNEEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGI 3726
Query: 265 NASLFTKKNLDSFIMLLVYVYDITLAG 291
+ +LF K++ ++ ++ +YV DI G
Sbjct: 3727 DKTLFVKQDAENLMIAQIYVDDIVFGG 3807
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 146 bits (368), Expect = 3e-35
Identities = 72/130 (55%), Positives = 91/130 (69%), Gaps = 1/130 (0%)
Frame = -2
Query: 123 VDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIR 182
V LPP P+GC WV VK G ++R K LVAKGY Q+ G+DY DTFS VAK+TT+R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 183 LVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLKP 241
L +A+A+I + LHQLD+ NAFLHGDL ED+YM PPG V+ + VCKL +SLYGLK
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 242 ASRKWYEKLT 251
+ R W+ K +
Sbjct: 46 SPRAWFGKFS 17
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 142 bits (358), Expect = 4e-34
Identities = 67/134 (50%), Positives = 90/134 (67%)
Frame = +3
Query: 85 LVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNV 144
L S T P + EA+ H W+Q M +E+ L+ GTW++V LPP +GC WV VK
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 145 DGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAF 204
+G ++R K LVAKGY Q+ G++Y DTFS V +TT+RL +A+A+I + LHQLD+ NAF
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 205 LHGDLHEDVYMAIP 218
LHGDL ED+YM P
Sbjct: 363 LHGDLEEDIYMEQP 404
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 127 bits (319), Expect = 1e-29
Identities = 67/146 (45%), Positives = 94/146 (63%), Gaps = 1/146 (0%)
Frame = -3
Query: 107 MQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGL 166
MQ EL ++ WK+V+ P + IG WV + K + G I R K LVAKGYNQ EG+
Sbjct: 455 MQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEGI 276
Query: 167 DYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVS-TSK 225
DY +T++ VA++ IR+++A SI L+Q+DV +AFL+G + E+VY+ PPG K
Sbjct: 275 DYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPDK 96
Query: 226 PNQVCKLSKSLYGLKPASRKWYEKLT 251
P V KL K+LYGLK A R WYE+++
Sbjct: 95 PTHVYKLQKALYGLKQAPRAWYERIS 18
>BM307983
Length = 406
Score = 110 bits (274), Expect = 2e-24
Identities = 58/133 (43%), Positives = 81/133 (60%), Gaps = 2/133 (1%)
Frame = +2
Query: 132 IGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASIN 191
+GC W+ VK+ D +++RYK LVAKGY Q G+DY +TF+ K
Sbjct: 2 VGCRWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQ 181
Query: 192 Y*F-LHQLDVNNAFLHGDLHEDVYMAIPPGVSTSK-PNQVCKLSKSLYGLKPASRKWYEK 249
+ + +HQ DV NAFLHG L E+VYM IPPG S N+VC+L K+LYGLK + R W+ +
Sbjct: 182 FGWEMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGR 361
Query: 250 LTCLPITNGYQQA 262
T ++ GY+Q+
Sbjct: 362 FTQAMLSLGYKQS 400
>AI959950
Length = 466
Score = 96.7 bits (239), Expect(2) = 2e-20
Identities = 53/131 (40%), Positives = 81/131 (61%), Gaps = 1/131 (0%)
Frame = -1
Query: 105 QVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIERYKVCLVAKGYNQIE 164
+ MQ EL + K+V LP K +G W+ K + DG + RYK LVAKGY+Q E
Sbjct: 394 KAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQE 215
Query: 165 GLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPGVSTS 224
G+DY TF+LVA++ I ++++ A+ + L+Q+DV +AFL+G + ++VY+ PPG
Sbjct: 214 GIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENE 35
Query: 225 KPNQ-VCKLSK 234
+Q V KL+K
Sbjct: 34 TLHQHVFKLNK 2
Score = 21.2 bits (43), Expect(2) = 2e-20
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = -2
Query: 84 SLVSYTEPKSYDEAIKHD 101
+L+ +PK DEAIK D
Sbjct: 456 ALIFEMKPKHIDEAIKDD 403
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 94.7 bits (234), Expect = 9e-20
Identities = 63/158 (39%), Positives = 86/158 (53%), Gaps = 2/158 (1%)
Frame = +1
Query: 146 GSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFL 205
G+I+++K LVAK Y Q+ G DY TFS VAK+ + L+ ++A + + L LD NAFL
Sbjct: 28 GTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFL 207
Query: 206 HGDLHEDVYMAIPPG--VSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQAT 263
HG L E+VYM P G N VC+L +S YGLK + R W L C Y
Sbjct: 208 HGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAW-PFLYC-GAAIWYDSHE 381
Query: 264 SNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIK 301
++ S+F + I L+VYV DI + G IT +K
Sbjct: 382 ADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK 495
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 93.6 bits (231), Expect = 2e-19
Identities = 51/111 (45%), Positives = 70/111 (62%)
Frame = +3
Query: 90 EPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIE 149
EPK+ EAI D W VMQ EL ++ WK+V+ P + IG WV + K + G I
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 150 RYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDV 200
R K LVAKGYNQ EG+DY +T++ VA++ IR+++A ASI L+Q+DV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 66.6 bits (161), Expect(2) = 2e-19
Identities = 33/94 (35%), Positives = 52/94 (55%)
Frame = +1
Query: 85 LVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNV 144
L + EP + +A+ W+Q MQ + L + T + LP I C WV ++K N+
Sbjct: 16 LFTTAEPSTVKQALISPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFRIKENL 195
Query: 145 DGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKV 178
G++ RY+ LVAKG++ G DY +TFS V ++
Sbjct: 196 YGTLNRYRSRLVAKGFHLKFGCDYSETFSPVIEL 297
Score = 47.4 bits (111), Expect(2) = 2e-19
Identities = 23/41 (56%), Positives = 30/41 (73%)
Frame = +3
Query: 180 TIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG 220
TIRL++ +A N+ L Q+D+NNAFLHG L E+VYM PG
Sbjct: 300 TIRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYMVQLPG 422
>CO983516
Length = 724
Score = 90.5 bits (223), Expect = 2e-18
Identities = 48/117 (41%), Positives = 78/117 (66%), Gaps = 1/117 (0%)
Frame = +2
Query: 172 FSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVC 230
F VA++ +IRL++ +A I L+Q+DV +AFL+G L+E+VY+ P G + + P+ V
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 231 KLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDI 287
+L K+LYGLK A R WYE+LT L GY++ + +LF K++ ++ ++ +YV DI
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDI 715
>BF595216
Length = 421
Score = 55.8 bits (133), Expect(2) = 4e-18
Identities = 26/48 (54%), Positives = 35/48 (72%)
Frame = +1
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALA 188
K N DG++ +YK LVAKG++Q G DY +TFSLV K T+RL++ LA
Sbjct: 124 KENSDGTVNKYKAKLVAKGFHQQYGTDYIETFSLVIKPITMRLLLTLA 267
Score = 53.9 bits (128), Expect(2) = 4e-18
Identities = 24/43 (55%), Positives = 32/43 (73%)
Frame = +3
Query: 195 LHQLDVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLY 237
+ QLDVNNAFL+G L E+VYM PPG ++ + VCKL K++Y
Sbjct: 291 IQQLDVNNAFLNGILQEEVYMQQPPGFDSTTKSLVCKLHKAIY 419
>BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Arabidopsis
thaliana}, partial (18%)
Length = 421
Score = 83.2 bits (204), Expect(2) = 5e-18
Identities = 48/83 (57%), Positives = 55/83 (65%)
Frame = -2
Query: 234 KSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDF 293
KSLYGLK ASRKWYEKLT L + GY Q+ S+ SLFT ++F LLVYV DI LAGD
Sbjct: 420 KSLYGLKQASRKWYEKLTNLLLKEGYIQSISDYSLFTLTKGNTFTALLVYVDDIILAGDS 241
Query: 294 LSEITFIKNALNQASKSKILVSL 316
+ E IKN L+ A K K L L
Sbjct: 240 IDEFDRIKNVLDLAFKIKNLGKL 172
Score = 26.2 bits (56), Expect(2) = 5e-18
Identities = 21/53 (39%), Positives = 24/53 (44%)
Frame = -3
Query: 308 SKSKILVSLNIS*ALKFVIPKVVFLYIIESIVWTYYKIHVSLVLNQSLHLLIH 360
SK KI VS N LK +I V +IVW +I L N HLL H
Sbjct: 197 SKLKIWVS*NTFLGLKLLILDWVLPSHRGNIVWICLRIQDYLGANLLAHLLTH 39
>AI855982
Length = 484
Score = 82.0 bits (201), Expect = 6e-16
Identities = 49/136 (36%), Positives = 74/136 (54%)
Frame = +2
Query: 83 MSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKH 142
M+ VS EPK+ EAI D W MQ EL ++ WK+V+ P + I WV + K
Sbjct: 77 MAFVSMIEPKNIKEAIVDDNWIIAMQEELNQFERNNVWKLVEKPDNYPVI*TKWVFRNKL 256
Query: 143 NVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNN 202
+ I +K LVA+GYNQ++GLDY T++ +A++ I + ++ I L+ +
Sbjct: 257 DEHRIIIIHKARLVAEGYNQVDGLDYEHTYASIARL*VIIMPLSYVYIMNSTLYHYACVS 436
Query: 203 AFLHGDLHEDVYMAIP 218
A LHG L +VY+ P
Sbjct: 437 ALLHGLLLHEVYVDQP 484
>TC232995
Length = 1009
Score = 58.9 bits (141), Expect = 6e-09
Identities = 29/71 (40%), Positives = 43/71 (59%), Gaps = 1/71 (1%)
Frame = +2
Query: 218 PPGVSTS-KPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDS 276
PPG S KPN V KL K+LYGLK A R WYE+L+ + + + + +LF K+ +
Sbjct: 11 PPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRKHND 190
Query: 277 FIMLLVYVYDI 287
+++ +YV DI
Sbjct: 191 ILLVQIYVDDI 223
>BM143109
Length = 415
Score = 56.6 bits (135), Expect = 3e-08
Identities = 28/70 (40%), Positives = 41/70 (58%)
Frame = +1
Query: 218 PPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSF 277
P ++ KPN V KL K LYGLK A R WYE L+ + G+ + + +LF K L+
Sbjct: 10 PVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLNDI 189
Query: 278 IMLLVYVYDI 287
+++ +YV DI
Sbjct: 190 LLVQIYVDDI 219
>BE474445
Length = 340
Score = 55.5 bits (132), Expect = 6e-08
Identities = 24/65 (36%), Positives = 34/65 (51%)
Frame = +1
Query: 72 TYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKP 131
T L SH + + EP++Y +AI+H W++ + EL + TW IV LP KP
Sbjct: 139 TQLFDSHKHYICQISENHEPQTYSQAIQHKPWQETISAELMAMKLNNTWTIVPLPQGKKP 318
Query: 132 IGCIW 136
I C W
Sbjct: 319 ISCKW 333
>CA801616 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberosum}, partial
(5%)
Length = 409
Score = 42.7 bits (99), Expect(2) = 1e-07
Identities = 21/48 (43%), Positives = 29/48 (59%)
Frame = +3
Query: 56 TNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDEAIKHDCW 103
T +S +PI NFLS+ LS S+S F SL S++ P + EA+ H W
Sbjct: 171 TRSSRNPHPIYNFLSYHRLSPSYSSFVFSLSSHSVPSNIHEALSHPGW 314
Score = 31.2 bits (69), Expect(2) = 1e-07
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +1
Query: 107 MQNELTTLDQTGTWKIVDLPPSSKPIG 133
M E+ L+ +GTW++ LPP K +G
Sbjct: 328 MIGEMQVLEHSGTWELFPLPPGKKAVG 408
>NP005470 reverse transcriptase
Length = 267
Score = 53.5 bits (127), Expect = 2e-07
Identities = 31/87 (35%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Frame = +1
Query: 203 AFLHGDLHEDVYMAIPPGVSTS-KPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQ 261
AFLHG L E++ M P G K V +L +SLYGLK + R+WY G+++
Sbjct: 7 AFLHGRLEENILMKQPEGFEVQGKERYVSQLQRSLYGLKQSPRQWYMSFDSFITNQGFKR 186
Query: 262 ATSNASLFTKKNLDS-FIMLLVYVYDI 287
+ + ++ K D I LL+YV D+
Sbjct: 187 SLYDCCVYHNKVEDGLMIYLLLYVDDM 267
>BM887535 similar to GP|14719283|gb Fourf gag/pol protein {Zea mays}, partial
(1%)
Length = 420
Score = 42.4 bits (98), Expect(2) = 6e-07
Identities = 22/67 (32%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Frame = +2
Query: 64 PISNFLSHTYLS---KSHSIFAMSLVSYTEPKSYDEAIKHDCWKQVMQNELTTLDQTGTW 120
P S H++ + H + L + P S EA+ H W+Q M +E L +GTW
Sbjct: 122 PSSGHSVHSFFGNRRRRHHMHYTCLSFVSIPNSPSEALSHLAWRQAMIDERCALQCSGTW 301
Query: 121 KIVDLPP 127
++V LPP
Sbjct: 302 ELVSLPP 322
Score = 29.3 bits (64), Expect(2) = 6e-07
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = +3
Query: 145 DGSIERYKVCLVAKGYNQ 162
DG+I+R+K LVAKGY Q
Sbjct: 348 DGTIDRFKAHLVAKGYTQ 401
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.336 0.146 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,300,862
Number of Sequences: 63676
Number of extensions: 433424
Number of successful extensions: 10704
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 6997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9833
length of query: 552
length of database: 12,639,632
effective HSP length: 102
effective length of query: 450
effective length of database: 6,144,680
effective search space: 2765106000
effective search space used: 2765106000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146555.15


