

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
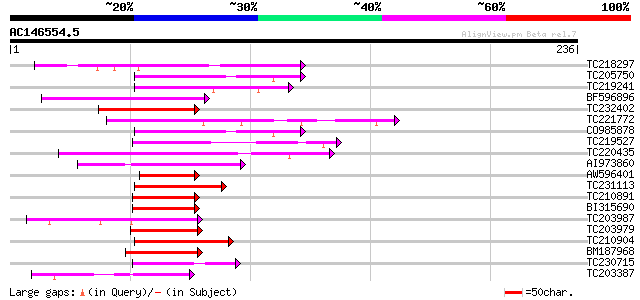
Query= AC146554.5 + phase: 0
(236 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218297 homologue to UP|ZFP4_ARATH (Q39263) Zinc finger protein... 62 3e-10
TC205750 homologue to UP|ZFP7_ARATH (Q39266) Zinc finger protein... 59 1e-09
TC219241 similar to UP|ZFP4_ARATH (Q39263) Zinc finger protein 4... 59 2e-09
BF596896 similar to SP|Q39261|ZFP2_ Zinc finger protein 2. [Mous... 59 2e-09
TC232402 homologue to UP|Q75I55 (Q75I55) Expressed protein, part... 58 4e-09
TC221772 homologue to UP|Q75I55 (Q75I55) Expressed protein, part... 58 4e-09
CO985878 52 2e-07
TC219527 homologue to UP|ZFP6_ARATH (Q39265) Zinc finger protein... 52 2e-07
TC220435 homologue to UP|ZFP6_ARATH (Q39265) Zinc finger protein... 50 9e-07
AI973860 49 2e-06
AW596401 49 2e-06
TC231113 homologue to UP|Q9FKA9 (Q9FKA9) Arabidopsis thaliana ge... 49 3e-06
TC210891 49 3e-06
BI315690 48 3e-06
TC203987 similar to UP|O22084 (O22084) ZPT2-12, partial (37%) 45 4e-05
TC203979 similar to UP|O22086 (O22086) ZPT2-14, partial (39%) 45 4e-05
TC210904 similar to UP|Q6YUF8 (Q6YUF8) C2H2-type zinc finger pro... 44 6e-05
BM187968 homologue to GP|2346974|dbj| ZPT2-12 {Petunia x hybrida... 43 1e-04
TC230715 similar to UP|Q39217 (Q39217) Zinc finger protein, part... 41 5e-04
TC203387 similar to UP|Q9XEU0 (Q9XEU0) Zinc-finger protein 1, pa... 40 7e-04
>TC218297 homologue to UP|ZFP4_ARATH (Q39263) Zinc finger protein 4, partial
(28%)
Length = 957
Score = 61.6 bits (148), Expect = 3e-10
Identities = 45/123 (36%), Positives = 61/123 (49%), Gaps = 10/123 (8%)
Frame = +2
Query: 11 DFLTLRLGTHTKSTSKPLENGGHSS---SFQNTLD---EPLPEQKVVE----FSCNYCDK 60
D ++L L + K++ E GG S SF +T + EP + FSCNYC +
Sbjct: 89 DAISLDLSLNFKNS----EPGGRDSIGFSFSSTSESSNEPASQTTAATIPRVFSCNYCQR 256
Query: 61 KFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYFRSANLHNPI 120
KF +SQALGGHQNAHKRER L K R + R+++ H FRS +
Sbjct: 257 KFFSSQALGGHQNAHKRERTLAK----RAMRMGFFSERYANLASLPLHGSFRSLGIKAHS 424
Query: 121 GVH 123
+H
Sbjct: 425 SLH 433
>TC205750 homologue to UP|ZFP7_ARATH (Q39266) Zinc finger protein 7, partial
(25%)
Length = 1464
Score = 59.3 bits (142), Expect = 1e-09
Identities = 35/72 (48%), Positives = 41/72 (56%), Gaps = 1/72 (1%)
Frame = +3
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGH-SYF 111
FSCNYC +KF +SQALGGHQNAHKRER + K R T R++S H S F
Sbjct: 612 FSCNYCRRKFYSSQALGGHQNAHKRERTMAKRAMRMGM----FTERYTSLASLPLHGSPF 779
Query: 112 RSANLHNPIGVH 123
RS L +H
Sbjct: 780 RSLGLEAHSAMH 815
>TC219241 similar to UP|ZFP4_ARATH (Q39263) Zinc finger protein 4, partial
(26%)
Length = 874
Score = 58.9 bits (141), Expect = 2e-09
Identities = 32/74 (43%), Positives = 42/74 (56%), Gaps = 8/74 (10%)
Frame = +3
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKK------MEDRRREEEMDLTLRFSSF--IH 104
FSCNYC +KF +SQALGGHQNAHKRER + K M R L L S+F +
Sbjct: 189 FSCNYCQRKFFSSQALGGHQNAHKRERTMAKRAMRMGMFAERYTSLASLPLHGSAFRSLG 368
Query: 105 YQGHSYFRSANLHN 118
+ H+ ++H+
Sbjct: 369 LEAHAAMHQGHVHH 410
>BF596896 similar to SP|Q39261|ZFP2_ Zinc finger protein 2. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (41%)
Length = 403
Score = 58.9 bits (141), Expect = 2e-09
Identities = 31/70 (44%), Positives = 39/70 (55%)
Frame = +2
Query: 14 TLRLGTHTKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQN 73
+L L H L+ SS ++ P+ + FSCNYC +KF +SQALGGHQN
Sbjct: 161 SLHLSQHDDENHLNLDLLLEPSSSSSSSPSPICSMEPRIFSCNYCQRKFYSSQALGGHQN 340
Query: 74 AHKRERVLKK 83
AHK ER L K
Sbjct: 341 AHKLERTLAK 370
>TC232402 homologue to UP|Q75I55 (Q75I55) Expressed protein, partial (18%)
Length = 1098
Score = 57.8 bits (138), Expect = 4e-09
Identities = 26/42 (61%), Positives = 30/42 (70%)
Frame = +2
Query: 38 QNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRER 79
QN+ PL FSCN+C +KF +SQALGGHQNAHKRER
Sbjct: 467 QNSSSRPLHSNNNKVFSCNFCMRKFYSSQALGGHQNAHKRER 592
>TC221772 homologue to UP|Q75I55 (Q75I55) Expressed protein, partial (14%)
Length = 770
Score = 57.8 bits (138), Expect = 4e-09
Identities = 47/140 (33%), Positives = 62/140 (43%), Gaps = 18/140 (12%)
Frame = +3
Query: 41 LDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRER-VLKKMEDRRREEEMDL---T 96
L +P FSCN+C +KF +SQALGGHQNAHKRER ++ + +R M T
Sbjct: 108 LSTAVPPATAKVFSCNFCMRKFFSSQALGGHQNAHKRERGAARRYQSQRSMAIMGFSMNT 287
Query: 97 LRFSSFIHYQGHSYFRSANLHNPI--GVHVTNAFPSWIGSPYGGYGGVYMPNTPLAT--- 151
L + Q HS +H P G+ V +F + Y + M TP T
Sbjct: 288 LTMCRSLGVQPHSL-----VHKPCRDGIMVAPSF-------HDAYARIGMAWTPFWTEDQ 431
Query: 152 ---------RYVMQMPNSPQ 162
R V + P SPQ
Sbjct: 432 ADMVWPGSFRLVPKQPQSPQ 491
>CO985878
Length = 769
Score = 52.4 bits (124), Expect = 2e-07
Identities = 32/72 (44%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Frame = -1
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGH-SYF 111
FSCNY KF +S LGGHQNAHKRER + K R R T R++S H S F
Sbjct: 649 FSCNYSCHKFFSSHVLGGHQNAHKRERTMAKRAMRMRM----FTERYTSLASLPLHGSTF 482
Query: 112 RSANLHNPIGVH 123
+S L +H
Sbjct: 481 QSLGLEARAAMH 446
>TC219527 homologue to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6, partial
(28%)
Length = 880
Score = 52.0 bits (123), Expect = 2e-07
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 6/93 (6%)
Frame = +3
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYF 111
++ C YC ++F+ SQALGGHQNAHK+ER L K Q F
Sbjct: 345 KYECQYCCREFANSQALGGHQNAHKKERQLLKR------------------AQMQAARGF 470
Query: 112 RSANLHNPIGVHVTNAFP------SWIGSPYGG 138
++ +HN I +++ P SW+ +P+GG
Sbjct: 471 VASQIHNTI---ISSLSPQSQPPLSWLCTPHGG 560
>TC220435 homologue to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6, partial
(21%)
Length = 820
Score = 50.1 bits (118), Expect = 9e-07
Identities = 38/117 (32%), Positives = 50/117 (42%), Gaps = 2/117 (1%)
Frame = +2
Query: 21 TKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERV 80
TK TS + S + P ++ C YC ++F+ SQALGGHQNAHK+ER
Sbjct: 155 TKVTSSTSPSSPSDSPDSSVAGAGPPSSADRKYECQYCCREFANSQALGGHQNAHKKERQ 334
Query: 81 LKKMEDRRREEEMDLTLRFSSFIHYQGHSYFRSAN--LHNPIGVHVTNAFPSWIGSP 135
K + + SF+ S F L P V V A PSW+ P
Sbjct: 335 QLKRAQLQASRNAAV-----SFVRNPMISAFAPPPHLLAPPGTVVVPAAPPSWVYVP 490
>AI973860
Length = 318
Score = 49.3 bits (116), Expect = 2e-06
Identities = 23/70 (32%), Positives = 41/70 (57%)
Frame = +2
Query: 29 ENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRR 88
+N S ++ D+ P Q V++SC++C + FS +QALGGH N H+R+R K
Sbjct: 77 QNPNQSEIRWSSDDQAGPGQ--VKYSCSFCQRGFSNAQALGGHMNIHRRDRAKLKQSAEE 250
Query: 89 REEEMDLTLR 98
+ +D++++
Sbjct: 251 KLLSLDISIK 280
>AW596401
Length = 429
Score = 48.9 bits (115), Expect = 2e-06
Identities = 20/25 (80%), Positives = 22/25 (88%)
Frame = +1
Query: 55 CNYCDKKFSTSQALGGHQNAHKRER 79
C+YC + F TSQALGGHQNAHKRER
Sbjct: 1 CHYCCRNFPTSQALGGHQNAHKRER 75
>TC231113 homologue to UP|Q9FKA9 (Q9FKA9) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24G6, partial (29%)
Length = 822
Score = 48.5 bits (114), Expect = 3e-06
Identities = 20/38 (52%), Positives = 26/38 (67%)
Frame = +1
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRRE 90
F C +C +KF +SQALGGHQNAHK+ER + + E
Sbjct: 214 FPCLFCSRKFYSSQALGGHQNAHKKERTAARKAKKASE 327
>TC210891
Length = 843
Score = 48.5 bits (114), Expect = 3e-06
Identities = 19/28 (67%), Positives = 23/28 (81%)
Frame = +1
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRER 79
+F C+YC + F SQALGGHQNAHK+ER
Sbjct: 127 KFRCHYCKRVFGNSQALGGHQNAHKKER 210
>BI315690
Length = 425
Score = 48.1 bits (113), Expect = 3e-06
Identities = 18/28 (64%), Positives = 24/28 (85%)
Frame = +2
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRER 79
++ C YC ++F+ SQALGGHQNAHK+ER
Sbjct: 230 KYECQYCCREFANSQALGGHQNAHKKER 313
>TC203987 similar to UP|O22084 (O22084) ZPT2-12, partial (37%)
Length = 742
Score = 44.7 bits (104), Expect = 4e-05
Identities = 29/90 (32%), Positives = 43/90 (47%), Gaps = 17/90 (18%)
Frame = +1
Query: 8 HVEDFLTL------RLGTHTKSTSKPLENGGHSSS--------FQNTLDEPLPEQK---V 50
H+EDFL L + + +N G S F ++ +QK
Sbjct: 52 HLEDFLILFGAKPLGYSNNMLGMKRHRDNEGSESLDLAKCLVLFSCPIESNKAQQKGFGA 231
Query: 51 VEFSCNYCDKKFSTSQALGGHQNAHKRERV 80
VEF C C +KFS+ QALGGH+ +HKR+++
Sbjct: 232 VEFECKTCSRKFSSFQALGGHRASHKRQKL 321
Score = 32.7 bits (73), Expect = 0.15
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Frame = +1
Query: 31 GGHSSSF--QNTLDEPLPEQ--------KVVEFSCNYCDKKFSTSQALGGHQNAHK 76
GGH +S Q E L EQ K C+ C +FS QALGGH H+
Sbjct: 286 GGHRASHKRQKLEGEELKEQAKTLSLWNKPKMHECSICGLEFSLGQALGGHMRKHR 453
>TC203979 similar to UP|O22086 (O22086) ZPT2-14, partial (39%)
Length = 820
Score = 44.7 bits (104), Expect = 4e-05
Identities = 17/30 (56%), Positives = 25/30 (82%)
Frame = +3
Query: 51 VEFSCNYCDKKFSTSQALGGHQNAHKRERV 80
VEF C C++KFS+ QALGGH+ +HKR+++
Sbjct: 306 VEFECKTCNRKFSSFQALGGHRASHKRQKL 395
Score = 32.7 bits (73), Expect = 0.15
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Frame = +3
Query: 31 GGHSSSF--QNTLDEPLPEQ--------KVVEFSCNYCDKKFSTSQALGGHQNAHK 76
GGH +S Q E L EQ K C+ C +FS QALGGH H+
Sbjct: 360 GGHRASHKRQKLEGEELKEQAKSLSLWNKPKMHECSICGLEFSLGQALGGHMRKHR 527
>TC210904 similar to UP|Q6YUF8 (Q6YUF8) C2H2-type zinc finger protein-like
protein, partial (20%)
Length = 989
Score = 43.9 bits (102), Expect = 6e-05
Identities = 16/41 (39%), Positives = 27/41 (65%)
Frame = +2
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEM 93
+SC++C ++F ++QALGGH N H+R+R K + E+
Sbjct: 485 YSCSFCRREFRSAQALGGHMNVHRRDRARLKQQPTSPHNEI 607
>BM187968 homologue to GP|2346974|dbj| ZPT2-12 {Petunia x hybrida}, partial
(29%)
Length = 424
Score = 42.7 bits (99), Expect = 1e-04
Identities = 16/32 (50%), Positives = 25/32 (78%)
Frame = +2
Query: 49 KVVEFSCNYCDKKFSTSQALGGHQNAHKRERV 80
+ V+F C C++KFS+ QALGGH+ +HKR ++
Sbjct: 203 EAVKFECKTCNRKFSSFQALGGHRASHKRSKL 298
Score = 28.1 bits (61), Expect = 3.6
Identities = 10/17 (58%), Positives = 13/17 (75%)
Frame = +2
Query: 55 CNYCDKKFSTSQALGGH 71
C+ C ++FS QALGGH
Sbjct: 365 CSICGQEFSLGQALGGH 415
>TC230715 similar to UP|Q39217 (Q39217) Zinc finger protein, partial (41%)
Length = 629
Score = 40.8 bits (94), Expect = 5e-04
Identities = 20/45 (44%), Positives = 27/45 (59%)
Frame = +2
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLT 96
EF C C++KFS+ QALGGH+ +HK+ K E +EE T
Sbjct: 161 EFECKTCNRKFSSFQALGGHRASHKK----PKFEAEELKEEAKKT 283
Score = 34.3 bits (77), Expect = 0.051
Identities = 18/52 (34%), Positives = 24/52 (45%), Gaps = 6/52 (11%)
Frame = +2
Query: 31 GGHSSSFQNT------LDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHK 76
GGH +S + L E + K C+ C +FS QALGGH H+
Sbjct: 212 GGHRASHKKPKFEAEELKEEAKKTKPKMHECSICGMEFSLGQALGGHMRKHR 367
>TC203387 similar to UP|Q9XEU0 (Q9XEU0) Zinc-finger protein 1, partial (51%)
Length = 1474
Score = 40.4 bits (93), Expect = 7e-04
Identities = 25/74 (33%), Positives = 41/74 (54%), Gaps = 6/74 (8%)
Frame = +3
Query: 10 EDFLTLRL------GTHTKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFS 63
E++L L L G + ST+KP + +S+ PL K+ + C+ C+K FS
Sbjct: 246 EEYLALCLIMLARGGAGSVSTAKPAVSDNNSA--------PLSAAKL-SYKCSVCNKAFS 398
Query: 64 TSQALGGHQNAHKR 77
+ QALGGH+ +H++
Sbjct: 399 SYQALGGHKASHRK 440
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/74 (32%), Positives = 31/74 (41%), Gaps = 11/74 (14%)
Frame = +3
Query: 13 LTLRLGTHTKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFS-----------CNYCDKK 61
L+ + K+ S GGH +S + E P V S C+ C K
Sbjct: 360 LSYKCSVCNKAFSSYQALGGHKASHRKLAGENHPTSSAVTTSSASNGGGRTHECSICHKT 539
Query: 62 FSTSQALGGHQNAH 75
FST QALGGH+ H
Sbjct: 540 FSTGQALGGHKRCH 581
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,103,395
Number of Sequences: 63676
Number of extensions: 189356
Number of successful extensions: 1092
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 1066
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1092
length of query: 236
length of database: 12,639,632
effective HSP length: 94
effective length of query: 142
effective length of database: 6,654,088
effective search space: 944880496
effective search space used: 944880496
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.5 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146554.5


