

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.15 - phase: 0
(880 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
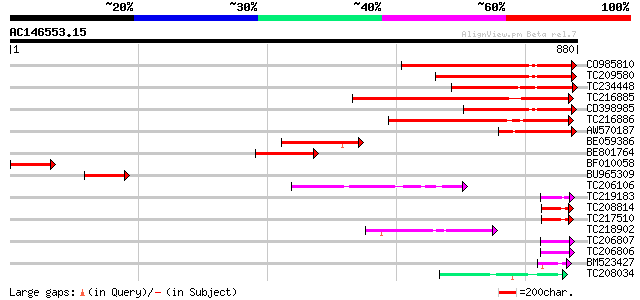
Score E
Sequences producing significant alignments: (bits) Value
CO985810 340 2e-93
TC209580 similar to PIR|T01393|T01393 apoptosis inhibitor homolo... 276 4e-74
TC234448 similar to PIR|T01393|T01393 apoptosis inhibitor homolo... 271 1e-72
TC216885 241 1e-63
CD398985 similar to PIR|T01393|T013 apoptosis inhibitor homolog ... 222 5e-58
TC216886 weakly similar to UP|BIR3_HUMAN (Q13489) Baculoviral IA... 207 2e-53
AW570187 similar to PIR|T01393|T013 apoptosis inhibitor homolog ... 130 3e-30
BE059386 124 2e-28
BE801764 100 5e-21
BF010058 99 8e-21
BU965309 67 3e-11
TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), ... 54 4e-07
TC219183 52 1e-06
TC208814 51 2e-06
TC217510 49 7e-06
TC218902 49 7e-06
TC206807 48 2e-05
TC206806 weakly similar to GB|BAB08380.1|9757771|AB005240 RING z... 48 2e-05
BM523427 48 2e-05
TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 48 2e-05
>CO985810
Length = 910
Score = 340 bits (872), Expect = 2e-93
Identities = 175/271 (64%), Positives = 213/271 (78%)
Frame = -3
Query: 609 GQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKS 668
GQVERAN RKLE+E AALRKE+EAAK+RA E+A + QEV +REKKTQMKFQSWE QKS
Sbjct: 902 GQVERANATXRKLEVEKAALRKEVEAAKIRATETAASCQEVXRREKKTQMKFQSWEKQKS 723
Query: 669 LLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEE 728
L QEEL EK KLA + +E +QA +Q EQ E + +Q AK EE + SSI+KEREQIEE
Sbjct: 722 LFQEELTIEKRKLAQLLQELEQARMQQEQVEGRWQQEAKAKEEFILQASSIKKEREQIEE 543
Query: 729 LARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGSYAGSFKD 788
++KE+ IKL+AE+ + Y+DDI KLEKEI+Q+R K+DSSKIAAL+ GIDG YA D
Sbjct: 542 SGKSKEDAIKLKAERNRQMYRDDIHKLEKEISQLRLKTDSSKIAALRMGIDGCYASKCLD 363
Query: 789 TKKGSGFEEPHTASISELVQKLNNFSMNGGGVKRERECVMCLSEEMSVVFLPCAHQVVCT 848
K G+ +EP + ISELV +++ + GGVKRE+ECVMCLSEEMSV+F+PCAHQVVC
Sbjct: 362 MKNGTAQKEPRASFISELV--IDHSAT--GGVKREQECVMCLSEEMSVLFMPCAHQVVCK 195
Query: 849 KCNELHEKQGMQDCPSCRSPIQERISVRYAR 879
CNELHEKQGMQDCPSCRSPIQ+RI+VR+ R
Sbjct: 194 TCNELHEKQGMQDCPSCRSPIQQRIAVRFPR 102
>TC209580 similar to PIR|T01393|T01393 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(7%)
Length = 803
Score = 276 bits (705), Expect = 4e-74
Identities = 143/219 (65%), Positives = 170/219 (77%)
Frame = +1
Query: 661 QSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIR 720
QSWE QKS +EEL EK KLA + E +QA VQ EQ E + +Q AK EEL+ SSIR
Sbjct: 1 QSWEKQKSFFKEELTIEKQKLAQLLHELEQARVQQEQVEGRWQQEAKAKEELILQASSIR 180
Query: 721 KEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDG 780
KEREQIEE ++KE+ IKL+AE+ L+ Y+DDIQKLEKEI+Q+R K+DSSKIA L+ GIDG
Sbjct: 181 KEREQIEESGKSKEDAIKLKAERNLQSYRDDIQKLEKEISQLRLKTDSSKIATLRMGIDG 360
Query: 781 SYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKRERECVMCLSEEMSVVFLP 840
YA F D K G+ +EP + ISELV +++ + G VKRERECVMCLSEEMSVVFLP
Sbjct: 361 CYARKFLDIKNGTAQKEPWASFISELV--IDHSAT--GSVKRERECVMCLSEEMSVVFLP 528
Query: 841 CAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYAR 879
CAHQVVCT CNELHEKQGMQDCPSCRSPIQ+RI+VR+ R
Sbjct: 529 CAHQVVCTPCNELHEKQGMQDCPSCRSPIQQRIAVRFPR 645
>TC234448 similar to PIR|T01393|T01393 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(7%)
Length = 666
Score = 271 bits (692), Expect = 1e-72
Identities = 139/195 (71%), Positives = 162/195 (82%)
Frame = +2
Query: 686 KESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKEL 745
+E +QA+VQ +Q EA+ +QAAK EELL SSIRKEREQIEE A++KE+ IKL+AE+ L
Sbjct: 2 QELEQAKVQQQQVEARWQQAAKAKEELLLQASSIRKEREQIEESAKSKEDMIKLKAEENL 181
Query: 746 RRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISE 805
RY+DDIQKLEKEIAQ+RQK+DSSKIAAL+RGIDG+Y SF D K +E ISE
Sbjct: 182 HRYRDDIQKLEKEIAQLRQKTDSSKIAALRRGIDGNYVSSFMDV-KSMALKESRATFISE 358
Query: 806 LVQKLNNFSMNGGGVKRERECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSC 865
+V LN++S+ GGVKRERECVMCLSEEMSVVFLPCAHQVVCT CN+LHEKQGMQDCPSC
Sbjct: 359 MVSNLNDYSLI-GGVKRERECVMCLSEEMSVVFLPCAHQVVCTTCNDLHEKQGMQDCPSC 535
Query: 866 RSPIQERISVRYART 880
RSPIQ RISVR+ART
Sbjct: 536 RSPIQRRISVRFART 580
>TC216885
Length = 1122
Score = 241 bits (614), Expect = 1e-63
Identities = 127/347 (36%), Positives = 214/347 (61%), Gaps = 4/347 (1%)
Frame = +3
Query: 533 MVPRVRELQNELQEWTEWANQKVMQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEE- 591
+V ++++L+ +++E +WA++K +QAA++LS D ELK + E+EE ++L KE EE
Sbjct: 12 LVNQIKDLEKQVKERKDWAHEKAIQAAKKLSSDLIELKKFKMEREENKKLPKETGAAEEL 191
Query: 592 --NTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQEV 649
TM +LSEMENAL K GQ+++A AVRKLE E A ++ E+EA+KL A ES T+ +V
Sbjct: 192 DNPTMMRLSEMENALRKTSGQMDQATAAVRKLEAEKAEIKAELEASKLSASESVTSCLQV 371
Query: 650 SKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKT 709
+KREKK K +WE QK + +++ EK K+ I +E Q + A++ E R++ K
Sbjct: 372 AKREKKCLKKLLTWEKQKVKIHQDISDEKQKILEIQEELAQIKQCAKETEVTRKEELKAK 551
Query: 710 EELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSS 769
EE L+++ R+ +E E + + ++L+ E + +R KDD+ +LE+EI++++ + S+
Sbjct: 552 EEALALIEEERRSKEAAEANHKRNLKALRLKIEIDFQRRKDDLLRLEQEISRLKAPARST 731
Query: 770 KIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNF-SMNGGGVKRERECVM 828
+ + S EP ++++L+ +L+N +G + +REC++
Sbjct: 732 TLPTSE-----------------SEDAEPQRETLAKLLLELDNVKDFSGKEINGDRECII 860
Query: 829 CLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISV 875
C +E+SV+FLPCAHQV+C +C + + K+G CP CR PI+ERI +
Sbjct: 861 CGKDEVSVIFLPCAHQVMCARCGKEYGKKGKAVCPCCRVPIEERIPI 1001
>CD398985 similar to PIR|T01393|T013 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana, partial (9%)
Length = 697
Score = 222 bits (566), Expect = 5e-58
Identities = 112/175 (64%), Positives = 139/175 (79%)
Frame = -1
Query: 705 AAKKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQ 764
+AK EE + SSI+KEREQIEE ++KE+ IKL+AE+ + Y+DDI KLEKEI+Q+R
Sbjct: 637 SAKAKEEFILQASSIKKEREQIEESGKSKEDAIKLKAERNRQMYRDDIHKLEKEISQLRL 458
Query: 765 KSDSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKRER 824
K+DSSKIAAL+ GIDG YA D K G+ +EP + ISELV +++ + GGVKRE+
Sbjct: 457 KTDSSKIAALRMGIDGCYASKCLDMKNGTAQKEPRASFISELV--IDHSAT--GGVKREQ 290
Query: 825 ECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYAR 879
ECVMCLSEEMSV+F+PCAHQVVC CNELHEKQGMQDCPSCRSPIQ+RI+VR+ R
Sbjct: 289 ECVMCLSEEMSVLFMPCAHQVVCKTCNELHEKQGMQDCPSCRSPIQQRIAVRFPR 125
>TC216886 weakly similar to UP|BIR3_HUMAN (Q13489) Baculoviral IAP
repeat-containing protein 3 (Inhibitor of apoptosis
protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF
signaling complex protein 1) (IAP homolog C) (Apoptosis
inhibitor 2) (API2), partial (4%)
Length = 1137
Score = 207 bits (527), Expect = 2e-53
Identities = 113/288 (39%), Positives = 176/288 (60%), Gaps = 1/288 (0%)
Frame = +1
Query: 589 LEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEMEAAKLRAVESATNFQE 648
LE+ TMK+LSEMENAL KA GQ++ N AVR+LE ENA ++ EMEA+KL A ES T E
Sbjct: 1 LEDTTMKRLSEMENALRKASGQLDLGNAAVRRLETENAEMKAEMEASKLSASESVTACLE 180
Query: 649 VSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEAKRRQAAKK 708
V+KREKK K +WE QK+ LQ+E+ EK K+ + Q ++ E K ++ K
Sbjct: 181 VAKREKKCLKKLLAWEKQKAKLQQEISDEKEKILKTKEILVQIRQCQKEAEVKWKEELKA 360
Query: 709 TEELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDS 768
EE L++V R +E E + K E ++L+ E + +R+KDD+ +LE+E+++++ + S
Sbjct: 361 KEEALALVEEERHCKEAAESNNKRKLEALRLKIEIDFQRHKDDLLRLEQELSRLKASAQS 540
Query: 769 SKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNF-SMNGGGVKRERECV 827
+++ + S D K +P +I+ L+Q+L+N + + REC+
Sbjct: 541 AELH------NQSSTSPTSDCKGA----KPQRETIARLLQELDNLEDFSEKEINSNRECI 690
Query: 828 MCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISV 875
+C+ +E+S+VFLPCAHQV+C C++ + ++G CP CR IQ+RI V
Sbjct: 691 VCMKDEVSIVFLPCAHQVMCASCSDEYGRKGKATCPCCRVQIQQRIRV 834
>AW570187 similar to PIR|T01393|T013 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana, partial (8%)
Length = 419
Score = 130 bits (326), Expect = 3e-30
Identities = 65/121 (53%), Positives = 84/121 (68%)
Frame = +1
Query: 759 IAQIRQKSDSSKIAALKRGIDGSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGG 818
+ ++ KSDS KIAAL+R +D SF TK + + S+ + + + G
Sbjct: 7 LVDLKLKSDSEKIAALRRCVDVRN-DSFSRTKSAPNMKGNKKSDTSQTLVSYQD-KLAAG 180
Query: 819 GVKRERECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVRYA 878
++RE+ECVMCLSEEMSVVFLPCAHQVVC +CNELHEKQGM++CPSCR+PIQ RI R+A
Sbjct: 181 SLRREQECVMCLSEEMSVVFLPCAHQVVCPECNELHEKQGMKECPSCRAPIQRRIHARFA 360
Query: 879 R 879
R
Sbjct: 361 R 363
>BE059386
Length = 406
Score = 124 bits (311), Expect = 2e-28
Identities = 72/135 (53%), Positives = 98/135 (72%), Gaps = 7/135 (5%)
Frame = +3
Query: 422 TQNNHNTHFSSNNGPSTPT-FSL-DSSDTISRAADSSSSEHEANLIPAVSSPPDALSATD 479
TQ++ + +FS N G ST T FSL +SSD++ R+ ++S + + AN IP S P +LSAT+
Sbjct: 3 TQDSISVNFSCNAGTSTSTAFSLVNSSDSVCRSTNTSFAINAANTIPVFSCPA-SLSATN 179
Query: 480 TDLSLSLSSKGNSSIAPICCSNKSHSSSCVGIPYD-----KSMRQWLPQDRKDELILKMV 534
TDLSLSLSSK S +C +N++ +SS +GI Y+ KS RQW+P D KDE+ILK++
Sbjct: 180 TDLSLSLSSKIKPSTESVCSNNEAPNSSYMGILYNNNNNNKSPRQWIPHDGKDEMILKLL 359
Query: 535 PRVRELQNELQEWTE 549
PRVRELQN+LQEWTE
Sbjct: 360 PRVRELQNQLQEWTE 404
>BE801764
Length = 308
Score = 99.8 bits (247), Expect = 5e-21
Identities = 58/100 (58%), Positives = 78/100 (78%), Gaps = 2/100 (2%)
Frame = +1
Query: 382 KLSGLSGLILDKKLKSVSESTAINLKSASINISKAVGIDVTQNNHNTHFSSNNGPSTPT- 440
+++GLSGL+LDKKLKSVSES+ INLKSAS+ ISKA+GID TQ+N N +FSSN G ST T
Sbjct: 10 RVNGLSGLVLDKKLKSVSESSTINLKSASLQISKAMGIDTTQDNINVNFSSNAGTSTSTA 189
Query: 441 FS-LDSSDTISRAADSSSSEHEANLIPAVSSPPDALSATD 479
FS +DSS+ + R+ ++S + + A+ IP S P +LSAT+
Sbjct: 190 FSPVDSSNAVCRSTNTSFAINAAHTIPLFSCPA-SLSATN 306
>BF010058
Length = 370
Score = 99.0 bits (245), Expect = 8e-21
Identities = 53/70 (75%), Positives = 57/70 (80%)
Frame = +2
Query: 1 MASLVVSGNSQMSSSVSVQEKGSRNKRKFRADPPLGESSKSISSLQHESLSYEFSAEKVE 60
MASLV SG+SQM+ SVSVQEKGSRNKRKFRADPPLGE +K I S QHESLS EFSAEK E
Sbjct: 161 MASLVASGSSQMAHSVSVQEKGSRNKRKFRADPPLGEPNKIIPSPQHESLSNEFSAEKFE 340
Query: 61 ITPCFGPVTA 70
IT G +A
Sbjct: 341 ITTGHGQASA 370
>BU965309
Length = 444
Score = 67.0 bits (162), Expect = 3e-11
Identities = 29/71 (40%), Positives = 46/71 (63%)
Frame = +1
Query: 116 ADWSDHTETQLQELVLSNLQTIFKSAIKKIVACGYTEDVATKAMLRPGICYGCKDTVSNI 175
+ W TE QL+ ++L N++ I+ + K+VA GY+E++A KA+L G CYG D +N+
Sbjct: 226 SSWVLCTEVQLETILLKNIEIIYNDTVPKLVALGYSEEIAVKAILYNGHCYGANDLATNV 405
Query: 176 VDNTLAFLRNG 186
+ N+LA L G
Sbjct: 406 LHNSLACLTTG 438
>TC206106 weakly similar to UP|Q7RK82 (Q7RK82) Maebl (Fragment), partial (7%)
Length = 1040
Score = 53.5 bits (127), Expect = 4e-07
Identities = 60/275 (21%), Positives = 116/275 (41%), Gaps = 2/275 (0%)
Frame = +2
Query: 438 TPTFSLDSSDTISRAADSSSSEHEANLIPAVSSPPDALSATDTDLSLSLSSKGNSSIAPI 497
TP + ++ A S E A A S D +S ++ + + + S +
Sbjct: 299 TPPTTQSEAEAEVEANAESIEEVVAEAEKAQSGVEDKISQSEAEAEAEVEANAKSIEEVV 478
Query: 498 CCSNKSHSSSCVGIPYDKSMRQWLPQDRKDELILKMVPRVRELQNELQEWTEWA--NQKV 555
+ K+ S + I S ++ + ++ + R+ +EL+ + A N ++
Sbjct: 479 AEAEKAQSGAEDKISQSVSFKE------ETNVVGDLPEAQRKALDELKRLVQEALNNHEL 640
Query: 556 MQAARRLSKDKAELKTLRQEKEEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERAN 615
K K ELK L +++EEV KKE + EE +++E + + A
Sbjct: 641 TAPKPEPEKKKTELKALEKKEEEVSEEKKEVEVTEEKKEAEVTE----------EKKEAE 790
Query: 616 TAVRKLEMENAALRKEMEAAKLRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELM 675
K E+E A +KE+E + + EV++ +K+T++ + +E +
Sbjct: 791 VTEEKKEVEVAEEKKEVEGTEEK------KEVEVAEEKKETEVIKEK--------KEVEV 928
Query: 676 TEKNKLAHISKESKQAEVQAEQFEAKRRQAAKKTE 710
TE+ K +++E K+AEV E+ E + K+ E
Sbjct: 929 TEEKKEVEVTEEKKEAEVIEEKKEVDATEEKKEAE 1033
Score = 36.2 bits (82), Expect = 0.064
Identities = 44/191 (23%), Positives = 82/191 (42%)
Frame = +2
Query: 577 EEVERLKKEKQCLEENTMKKLSEMENALGKAGGQVERANTAVRKLEMENAALRKEMEAAK 636
E +E + E + + K+S+ E +A +VE AN + +E A K A+
Sbjct: 350 ESIEEVVAEAEKAQSGVEDKISQSE---AEAEAEVE-ANA--KSIEEVVAEAEKAQSGAE 511
Query: 637 LRAVESATNFQEVSKREKKTQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAE 696
+ +S + +E + + + ++ + K L+QE L + E K+ E++A
Sbjct: 512 DKISQSVSFKEETNVVGDLPEAQRKALDELKRLVQEALNNHELTAPKPEPEKKKTELKAL 691
Query: 697 QFEAKRRQAAKKTEELLSMVSSIRKEREQIEELARTKEERIKLEAEKELRRYKDDIQKLE 756
+ KK EE VS +KE E EE + K EAE + + ++ + +
Sbjct: 692 E---------KKEEE----VSEEKKEVEVTEEKKEAEVTEEKKEAEVTEEKKEVEVAEEK 832
Query: 757 KEIAQIRQKSD 767
KE+ +K +
Sbjct: 833 KEVEGTEEKKE 865
>TC219183
Length = 742
Score = 51.6 bits (122), Expect = 1e-06
Identities = 20/53 (37%), Positives = 30/53 (55%)
Frame = +1
Query: 824 RECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVR 876
+ECV+C++E LPC H +C++C H Q CP CR I+E I ++
Sbjct: 169 KECVICMTEPKDTAVLPCRHMCMCSECANAHRLQS-NKCPICRQSIEELIEIK 324
>TC208814
Length = 638
Score = 51.2 bits (121), Expect = 2e-06
Identities = 21/50 (42%), Positives = 32/50 (64%)
Frame = +1
Query: 826 CVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISV 875
C +C E+++VV LPC H ++C+ C E ++ CP CR PI+ER+ V
Sbjct: 343 CRICFEEQINVVLLPCRHHILCSTCCEKCKR-----CPVCRGPIEERMPV 477
>TC217510
Length = 791
Score = 49.3 bits (116), Expect = 7e-06
Identities = 22/50 (44%), Positives = 31/50 (62%)
Frame = +1
Query: 826 CVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISV 875
C +C E++VV LPC H+V+C+ C+E +K CP CR I ER+ V
Sbjct: 334 CRVCFEGEINVVLLPCRHRVLCSTCSEKCKK-----CPICRDSIAERLPV 468
>TC218902
Length = 1098
Score = 49.3 bits (116), Expect = 7e-06
Identities = 61/226 (26%), Positives = 97/226 (41%), Gaps = 21/226 (9%)
Frame = +2
Query: 553 QKVMQAARRLSKDKAELKTLRQE-----------KEEVERLKKEKQCLEENTMKK---LS 598
Q+ A+ ++ +AEL+ R E KE++ L K+ Q E T +
Sbjct: 23 QREGMASVAVASLEAELEKTRSEIALVQMKEKEAKEKMTELPKKLQLTAEETNQANLLAQ 202
Query: 599 EMENALGKAGGQVERANTAVRKLEMENAALRKEMEAAKLR---AVESATNFQEVSKREKK 655
L K + E+A V LE A +KE+EAAK A+ + QE K
Sbjct: 203 AAREELQKVKAEAEQAKAGVSTLESRLLAAQKEIEAAKASENLAIAAIKALQESESTRSK 382
Query: 656 TQMKFQSWENQKSLLQEELMTEKNKLAHISKESKQAEVQAEQFEA-KRRQAAKKTEELLS 714
++ N +L EE E +K AH ++E V A E K +++ K E L
Sbjct: 383 NEV---DPSNGVTLSLEEYY-ELSKRAHEAEERANMRVAAANSEIDKVKESELKAFEKLD 550
Query: 715 MVSSIRKEREQIEELARTKEERI---KLEAEKELRRYKDDIQKLEK 757
V+ R + +LA K E+ KL E+ELR+++ + ++ K
Sbjct: 551 EVNREIAARRESLKLAMEKAEKAKEGKLGVEQELRKWRAESEQRRK 688
>TC206807
Length = 701
Score = 48.1 bits (113), Expect = 2e-05
Identities = 19/53 (35%), Positives = 30/53 (55%)
Frame = +2
Query: 824 RECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVR 876
+ECV+CLSE LPC H +C+ C ++ Q CP CR P++ + ++
Sbjct: 275 KECVICLSEPRDTTVLPCRHMCMCSGCAKVLRFQ-TNRCPICRQPVERLLEIK 430
>TC206806 weakly similar to GB|BAB08380.1|9757771|AB005240 RING zinc finger
protein-like {Arabidopsis thaliana;} , partial (66%)
Length = 1433
Score = 48.1 bits (113), Expect = 2e-05
Identities = 19/53 (35%), Positives = 30/53 (55%)
Frame = +2
Query: 824 RECVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQERISVR 876
+ECV+CLSE LPC H +C+ C ++ Q CP CR P++ + ++
Sbjct: 1001 KECVICLSEPRDTTVLPCRHMCMCSGCAKVLRFQ-TNRCPICRQPVERLLEIK 1156
>BM523427
Length = 419
Score = 48.1 bits (113), Expect = 2e-05
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Frame = +1
Query: 819 GVKRERE----CVMCLSEEMSVVFLPCAHQVVCTKCNELHEKQGMQDCPSCRSPIQE 871
GVK++R CV+CL +E + VF+PC H CT C+ + +CP CR I++
Sbjct: 40 GVKKDRLMPDLCVICLEQEYNAVFVPCGHMCCCTTCS-----SHLTNCPLCRRQIEK 195
>TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1183
Score = 47.8 bits (112), Expect = 2e-05
Identities = 51/206 (24%), Positives = 82/206 (39%), Gaps = 8/206 (3%)
Frame = +3
Query: 668 SLLQEELMTE-KNKLAHISKESKQAEVQAEQFEAKRRQAAKKTEELLSMVSSIRKEREQI 726
SLL E L ++ K + I + + E Q + A++RQ +T + S +R+ RE+
Sbjct: 312 SLLSEGLSSQIKQQRDEIDQFLQAQEEQLRRALAEKRQRHYRTLLRAAEESVLRRLREKE 491
Query: 727 EELARTKEERIKLEAEKELRRYKDDIQKLEKEIAQIRQKSDSSKIAALKRGI-------D 779
EL + +LEA +E ++ Q R K+ + AAL+ + D
Sbjct: 492 AELEKATRHNAELEARATQL-------SVEAQLWQARAKAQEATAAALQAQLQQAMMIGD 650
Query: 780 GSYAGSFKDTKKGSGFEEPHTASISELVQKLNNFSMNGGGVKRERECVMCLSEEMSVVFL 839
G G + G G E+ +A + V +C C SVV L
Sbjct: 651 GENGGGGGLSCAGGGAEDAESAYVDP------------DRVGPTPKCRGCAKRVASVVVL 794
Query: 840 PCAHQVVCTKCNELHEKQGMQDCPSC 865
PC H +C +C+ + CP C
Sbjct: 795 PCRHLCICAECD-----THFRACPVC 857
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.125 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,039,321
Number of Sequences: 63676
Number of extensions: 500193
Number of successful extensions: 3013
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 2881
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2960
length of query: 880
length of database: 12,639,632
effective HSP length: 106
effective length of query: 774
effective length of database: 5,889,976
effective search space: 4558841424
effective search space used: 4558841424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146553.15


