

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.13 + phase: 0
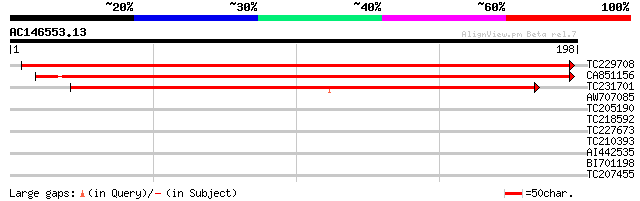
(198 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229708 weakly similar to UP|Q9LVF1 (Q9LVF1) Emb|CAB45066.1 (AT... 303 3e-83
CA851156 268 9e-73
TC231701 147 4e-36
AW707085 similar to GP|4680205|gb|A H beta 58 homolog {Sorghum b... 28 2.1
TC205190 28 2.8
TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, pa... 27 6.2
TC227673 similar to UP|Q9LKG1 (Q9LKG1) Histone deacetylase, part... 27 6.2
TC210393 weakly similar to GB|AAF25189.1|6694227|AF187147 drebri... 27 8.0
AI442535 27 8.0
BI701198 similar to GP|6739274|gb|A abscisic acid responsive ele... 27 8.0
TC207455 similar to UP|Q9XHE5 (Q9XHE5) Microtubule-associated pr... 27 8.0
>TC229708 weakly similar to UP|Q9LVF1 (Q9LVF1) Emb|CAB45066.1
(AT3g21550/MIL23_11), partial (88%)
Length = 868
Score = 303 bits (776), Expect = 3e-83
Identities = 140/193 (72%), Positives = 167/193 (85%)
Frame = +2
Query: 5 ASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG 64
AS + T ++ +S M+ KTFSG+GNLVKLLPTGTVFLFQ+L PVVTN+GHC+T++KYL+G
Sbjct: 20 ASKTATGSSSSSSRMSTKTFSGLGNLVKLLPTGTVFLFQFLIPVVTNSGHCTTLHKYLTG 199
Query: 65 ILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHA 124
LV+C FNCAF SFTDSYTGSDG+RHY +VT GLWPSP S+SV+LSAYKLRFGDFVHA
Sbjct: 200 SFLVVCAFNCAFASFTDSYTGSDGERHYALVTAKGLWPSPASESVNLSAYKLRFGDFVHA 379
Query: 125 FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
F S++VFAVLGLLDTN V CFYP FES+EKILMQV+PPVIG V+ VF++FP+ RHGIGY
Sbjct: 380 FFSLVVFAVLGLLDTNTVRCFYPAFESAEKILMQVVPPVIGAVASTVFVMFPNNRHGIGY 559
Query: 185 PTSSDTNDTSEKT 197
PTSSD+NDT K+
Sbjct: 560 PTSSDSNDTQHKS 598
>CA851156
Length = 688
Score = 268 bits (686), Expect = 9e-73
Identities = 126/188 (67%), Positives = 156/188 (82%)
Frame = +3
Query: 10 TSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVI 69
+S N+G +T T S G+L+KLLPTGTVF+FQ+++PV+TN+G C+ +K+L ILLV+
Sbjct: 102 SSQNRGTG-VTNTTLSAFGSLIKLLPTGTVFVFQFVNPVLTNSGDCNATSKWLCSILLVL 278
Query: 70 CGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHAFLSVI 129
CGF+CAF+SFTDSYTGSD QRHYGIVT GLWPSP S++VDLS YKL+FGD VHA LS+
Sbjct: 279 CGFSCAFSSFTDSYTGSDNQRHYGIVTTKGLWPSPASNTVDLSTYKLKFGDLVHAVLSLS 458
Query: 130 VFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSD 189
VFAVLGLLDTN VHCFYP FES++K L+QVLP IGV +G +FMIFP+ RHGIGYP +SD
Sbjct: 459 VFAVLGLLDTNTVHCFYPGFESTQKRLLQVLPTAIGVFAGGLFMIFPNDRHGIGYPLTSD 638
Query: 190 TNDTSEKT 197
+NDTS K+
Sbjct: 639 SNDTSPKS 662
>TC231701
Length = 764
Score = 147 bits (370), Expect = 4e-36
Identities = 70/167 (41%), Positives = 104/167 (61%), Gaps = 3/167 (1%)
Frame = +3
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTD 81
+TF +L LLPTGTV FQ LSP+ TN G+C +++K ++ L+ +CG +C + TD
Sbjct: 264 QTFQSTAHLGNLLPTGTVLAFQLLSPIFTNVGNCDSVSKAMTAALVSLCGASCFMSCLTD 443
Query: 82 SYTGSDGQRHYGIVTMNGLWPSPGSDSVD---LSAYKLRFGDFVHAFLSVIVFAVLGLLD 138
S+ S G YG T+ GLW GS ++ + Y+L+ DF+HA +SV+VFA + L D
Sbjct: 444 SFRDSKGSICYGFATLRGLWVIDGSTTLPPQLAAKYRLKLIDFMHAVMSVLVFAAIALFD 623
Query: 139 TNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYP 185
NVV+CF+P + + ++ LP IGV+ F+ FP+ RHGIG+P
Sbjct: 624 QNVVNCFFPAPSTETQEILTALPVGIGVLGSMFFVAFPTQRHGIGFP 764
>AW707085 similar to GP|4680205|gb|A H beta 58 homolog {Sorghum bicolor},
partial (2%)
Length = 388
Score = 28.5 bits (62), Expect = 2.1
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Frame = -1
Query: 32 KLLPTGTVFLFQYLSPVVTNNGHCSTINKY-LSGILLVICGFN 73
+++ + + Y S + T NGH I+KY +S I+ +I FN
Sbjct: 265 RIVSNNKMLVLHYDSSIATVNGHIPYISKYNISTIICIIWDFN 137
>TC205190
Length = 812
Score = 28.1 bits (61), Expect = 2.8
Identities = 27/80 (33%), Positives = 37/80 (45%)
Frame = -1
Query: 2 SEKASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKY 61
SEK +S +TSTNKG T K L LL T L+ + +N H ST
Sbjct: 638 SEK-TSKRTSTNKGPXXKTKKKKQN-NYLAFLLYIYTSHRHSSLAS-INHNHHTST---- 480
Query: 62 LSGILLVICGFNCAFTSFTD 81
+L + C +C+F +F D
Sbjct: 479 --KVLSIGCESHCSFLAFID 426
>TC218592 weakly similar to UP|Q9LP32 (Q9LP32) T9L6.1 protein, partial (37%)
Length = 715
Score = 26.9 bits (58), Expect = 6.2
Identities = 9/35 (25%), Positives = 20/35 (56%)
Frame = +3
Query: 114 YKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPK 148
+K + + H F+ +F+V ++ N+V+ + PK
Sbjct: 579 FKFKINESSHGFIYGNIFSVASVIQANIVYLYGPK 683
>TC227673 similar to UP|Q9LKG1 (Q9LKG1) Histone deacetylase, partial (88%)
Length = 1476
Score = 26.9 bits (58), Expect = 6.2
Identities = 12/48 (25%), Positives = 26/48 (54%)
Frame = +1
Query: 117 RFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVI 164
RF FL ++FA+L ++ N++H +P +++M+ + P +
Sbjct: 706 RFISLGITFLEQVIFAILDMVKGNIIH*MFPWM---MELMMRAISPCL 840
>TC210393 weakly similar to GB|AAF25189.1|6694227|AF187147 drebrin A {Mus
musculus;} , partial (4%)
Length = 784
Score = 26.6 bits (57), Expect = 8.0
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 10/72 (13%)
Frame = -1
Query: 26 GVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG----------ILLVICGFNCA 75
G +L++LLPT V F S + HC + ++YLS I L C F+
Sbjct: 217 GHSSLMQLLPTSGVAAFTVASTKESVCCHCCSRSRYLSSSCSIC*RSXDIELCSCSFSFL 38
Query: 76 FTSFTDSYTGSD 87
+S S+ +D
Sbjct: 37 SSSLILSFKMAD 2
>AI442535
Length = 420
Score = 26.6 bits (57), Expect = 8.0
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = -3
Query: 141 VVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPS 177
VVHC E +K+ + L V+ V GAV ++F S
Sbjct: 256 VVHCSTNA*EQLQKLHLLALEKVVVAVVGAVVLVFGS 146
>BI701198 similar to GP|6739274|gb|A abscisic acid responsive
elements-binding factor {Arabidopsis thaliana}, partial
(7%)
Length = 421
Score = 26.6 bits (57), Expect = 8.0
Identities = 14/37 (37%), Positives = 20/37 (53%)
Frame = -3
Query: 141 VVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPS 177
VVHC E +K+ + L V+ V GAV ++F S
Sbjct: 125 VVHCSTNA*EQLQKLHLLALEKVVVAVVGAVVLVFGS 15
>TC207455 similar to UP|Q9XHE5 (Q9XHE5) Microtubule-associated protein,
partial (11%)
Length = 975
Score = 26.6 bits (57), Expect = 8.0
Identities = 15/57 (26%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = +3
Query: 6 SSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLF---QYLSPVVTNNGHCSTIN 59
SS+ + N ++T+ T + N ++P+ T + + P T NGHCS+++
Sbjct: 453 SSTAHNCNNQSNTIPSSTANCCNNQSNIIPSSTANNCNNQRNIIPSSTANGHCSSVS 623
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,759,237
Number of Sequences: 63676
Number of extensions: 122583
Number of successful extensions: 615
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 614
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 614
length of query: 198
length of database: 12,639,632
effective HSP length: 92
effective length of query: 106
effective length of database: 6,781,440
effective search space: 718832640
effective search space used: 718832640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146553.13


