

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.9 - phase: 0 /pseudo
(454 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
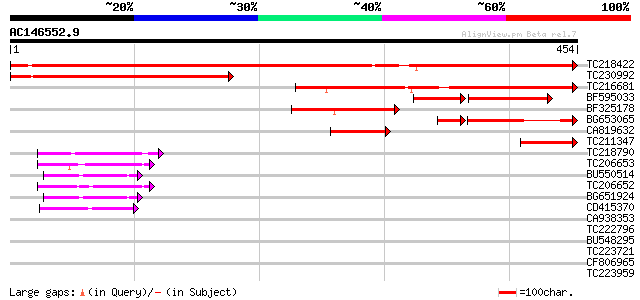
Score E
Sequences producing significant alignments: (bits) Value
TC218422 similar to UP|Q9FUY4 (Q9FUY4) Protein kinase AKINbetaga... 666 0.0
TC230992 similar to UP|Q944A6 (Q944A6) At1g09020/F7G19_11, parti... 299 2e-81
TC216681 similar to UP|Q944A6 (Q944A6) At1g09020/F7G19_11, parti... 275 3e-74
BF595033 similar to GP|16612255|gb| At1g09020/F7G19_11 {Arabidop... 112 8e-39
BF325178 weakly similar to GP|29887975|gb| AKIN betagamma {Medic... 112 3e-25
BG653065 similar to GP|11139548|gb protein kinase AKINbetagamma-... 92 9e-23
CA819632 69 4e-12
TC211347 similar to UP|Q84PE0 (Q84PE0) AKIN betagamma, partial (... 60 2e-09
TC218790 similar to UP|Q7XYX7 (Q7XYX7) AKIN beta2, partial (89%) 52 4e-07
TC206653 similar to UP|Q7XYX8 (Q7XYX8) AKIN beta1, partial (97%) 52 6e-07
BU550514 weakly similar to GP|8778877|gb| T7N9.13 {Arabidopsis t... 52 7e-07
TC206652 similar to UP|Q7XYX8 (Q7XYX8) AKIN beta1, partial (96%) 50 3e-06
BG651924 weakly similar to GP|8778877|gb| T7N9.13 {Arabidopsis t... 50 3e-06
CD415370 45 5e-05
CA938353 similar to GP|15010688|gb AT5g39790/MKM21_80 {Arabidops... 40 0.002
TC222796 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, parti... 40 0.002
BU548295 similar to GP|15010688|gb| AT5g39790/MKM21_80 {Arabidop... 37 0.024
TC223721 similar to UP|Q9LFY0 (Q9LFY0) T7N9.13, partial (10%) 37 0.024
CF806965 31 1.3
TC223959 similar to UP|Q9LFY0 (Q9LFY0) T7N9.13, partial (5%) 31 1.3
>TC218422 similar to UP|Q9FUY4 (Q9FUY4) Protein kinase AKINbetagamma-2,
partial (81%)
Length = 1968
Score = 666 bits (1718), Expect = 0.0
Identities = 349/460 (75%), Positives = 383/460 (82%), Gaps = 6/460 (1%)
Frame = +1
Query: 1 MFSPSMDSARDVGGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTV 60
MFSPSMDSARD GV AGTVLIP+RFVWPYGGR+VYLSGSFTRWSELLQMSPVEGCPTV
Sbjct: 76 MFSPSMDSARDASGV--AGTVLIPMRFVWPYGGRSVYLSGSFTRWSELLQMSPVEGCPTV 249
Query: 61 FQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATDP-FVPVLPPDIVSG 119
FQVIH+L PG+HQYKFFVDGEWRHD+H P ++G+YGIVNTV LATDP VPVL P+IVSG
Sbjct: 250 FQVIHSLIPGHHQYKFFVDGEWRHDDHQPCVSGEYGIVNTVSLATDPNIVPVLTPEIVSG 429
Query: 120 SNMDVDNETFQRVVRLTDGTLSEVM-PRISDVDVQTSRQRISTYLSMRTAYELLPESGKV 178
SNMDVDNE F+R+VRLTDGTLS V+ PRISDVD+QTSRQRIS +LSM TAYELLPESGKV
Sbjct: 430 SNMDVDNEAFRRMVRLTDGTLSNVLLPRISDVDIQTSRQRISAFLSMSTAYELLPESGKV 609
Query: 179 VTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEE 238
VTLDVDLPVKQAFHILHEQGIP+APLWD CKGQFVGVLS LDFILILRELGNHGSNLTEE
Sbjct: 610 VTLDVDLPVKQAFHILHEQGIPIAPLWDICKGQFVGVLSALDFILILRELGNHGSNLTEE 789
Query: 239 ELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQ 298
ELETHTISAWK GKWT F++ FI AGP DNLK++A+KILQNGISTVPIIHS DGSFPQ
Sbjct: 790 ELETHTISAWKGGKWTGFTQCFIRAGPYDNLKEIAVKILQNGISTVPIIHSE--DGSFPQ 963
Query: 299 LLHLASLSGILRCILGVVLVHCPYFN--FQFVQFLWAHGCP-KLGRQIAGL*QH*DQMLL 355
LLHLASLSGIL+CI C YF + L C +G + + + + L
Sbjct: 964 LLHLASLSGILKCI-------CRYFRNCSSSLPILQLPICAIPVGTWVPKIGESNRRPLA 1122
Query: 356 LLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTV 414
+L+P N+ AQVSSIPIVD+SDSLLDIYCRSDITALAKDR YTHINLDEMTV
Sbjct: 1123MLRPNASLTSALNLLVQAQVSSIPIVDDSDSLLDIYCRSDITALAKDRTYTHINLDEMTV 1302
Query: 415 HQALQLSQDAFNPNESRSQRCQMCLRTDSLHKVMERLANP 454
HQALQL QD++N E QRCQMCLRTDSLHKVMERLA+P
Sbjct: 1303HQALQLGQDSYNTYELSCQRCQMCLRTDSLHKVMERLASP 1422
>TC230992 similar to UP|Q944A6 (Q944A6) At1g09020/F7G19_11, partial (24%)
Length = 826
Score = 299 bits (766), Expect = 2e-81
Identities = 144/180 (80%), Positives = 161/180 (89%), Gaps = 1/180 (0%)
Frame = +3
Query: 1 MFSPSMDSARDVGGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTV 60
MF SMDSARD G VA GTVLIP+RFVWPYGGR+V+LSGSFTRW ELL MSPVEGCPTV
Sbjct: 285 MFGQSMDSARDAAGGVA-GTVLIPMRFVWPYGGRSVFLSGSFTRWLELLPMSPVEGCPTV 461
Query: 61 FQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATDP-FVPVLPPDIVSG 119
FQVI+NL PGYHQYKFFVDGEWRHDEH P++ G+YGIVNTVLLATDP ++PVLPPD+ SG
Sbjct: 462 FQVIYNLPPGYHQYKFFVDGEWRHDEHQPYVPGEYGIVNTVLLATDPNYIPVLPPDVASG 641
Query: 120 SNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGKVV 179
++MDVDN+ F+R+VRLTDGTLSEV+PRISD DVQ SRQRIS +LS TAYELLPESGKVV
Sbjct: 642 NSMDVDNDAFRRMVRLTDGTLSEVLPRISDTDVQISRQRISAFLSSHTAYELLPESGKVV 821
>TC216681 similar to UP|Q944A6 (Q944A6) At1g09020/F7G19_11, partial (50%)
Length = 1328
Score = 275 bits (703), Expect = 3e-74
Identities = 155/240 (64%), Positives = 170/240 (70%), Gaps = 15/240 (6%)
Frame = +2
Query: 230 NHGSNLTEEELETHTISAWKEGK----------WTLFSRRFIHAGPSDNLKDVALKILQN 279
NHGSNLTEEELETHTISAWKEGK T FSRRFIHAGP DNLKD+A+KILQ
Sbjct: 2 NHGSNLTEEELETHTISAWKEGKSYLNRQNNGHGTTFSRRFIHAGPYDNLKDIAMKILQK 181
Query: 280 GISTVPIIHSSSADGSFPQLLHLASLSGILRCILGVVLVHC----PYFNFQFVQFLWAHG 335
+STVPIIHSSS D SFPQLLHLASLSGIL+CI HC P
Sbjct: 182 EVSTVPIIHSSSEDASFPQLLHLASLSGILKCICRYFR-HCSSSLPVLQLPICAIPVGTW 358
Query: 336 CPKLGRQIAGL*QH*DQMLLLLQP*IY*FK-FNIFFAAQVSSIPIVDESDSLLDIYCRSD 394
PK+G + L +L+P N+ AQVSSIPIVD++DSLLDIYCRSD
Sbjct: 359 VPKIGESNR-------RPLAMLRPTASLASALNLLVQAQVSSIPIVDDNDSLLDIYCRSD 517
Query: 395 ITALAKDRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQMCLRTDSLHKVMERLANP 454
ITALAK+RAYTHINLDEMTVHQALQL QDA++P E RSQRCQMCLR+D LHKVMERLANP
Sbjct: 518 ITALAKNRAYTHINLDEMTVHQALQLGQDAYSPYELRSQRCQMCLRSDPLHKVMERLANP 697
>BF595033 similar to GP|16612255|gb| At1g09020/F7G19_11 {Arabidopsis
thaliana}, partial (11%)
Length = 413
Score = 112 bits (280), Expect(2) = 8e-39
Identities = 55/67 (82%), Positives = 62/67 (92%)
Frame = +3
Query: 368 IFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQDAFNP 427
++F AQVSSIPIVD++DSLLDIYCRSDITALAK+RAYTHINLDEMTVHQALQL DA++P
Sbjct: 213 LYF*AQVSSIPIVDDNDSLLDIYCRSDITALAKNRAYTHINLDEMTVHQALQLGXDAYSP 392
Query: 428 NESRSQR 434
E RSQR
Sbjct: 393 YELRSQR 413
Score = 66.6 bits (161), Expect(2) = 8e-39
Identities = 34/42 (80%), Positives = 35/42 (82%)
Frame = +1
Query: 324 NFQFVQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQP*IY*FK 365
NFQ VQ+LWA GCPKLG QI GL*Q *DQ LLL QP*IY*FK
Sbjct: 4 NFQSVQYLWARGCPKLGNQIGGL*QC*DQPLLLRQP*IY*FK 129
>BF325178 weakly similar to GP|29887975|gb| AKIN betagamma {Medicago
truncatula}, partial (14%)
Length = 361
Score = 112 bits (281), Expect = 3e-25
Identities = 59/97 (60%), Positives = 70/97 (71%), Gaps = 10/97 (10%)
Frame = +2
Query: 226 RELGNHGSNLTEEELETHTISAWKEGKWTLFSR----------RFIHAGPSDNLKDVALK 275
+ELGNHGSNLT+E+LETHTI+AWKEGK+ F +FIH P LK++ALK
Sbjct: 2 KELGNHGSNLTQEQLETHTIAAWKEGKFQQFRTLDXNGGSYP*QFIHTEPHKYLKNIALK 181
Query: 276 ILQNGISTVPIIHSSSADGSFPQLLHLASLSGILRCI 312
+LQN TVPIIHSSS D ++ QLLHL SLSGILR I
Sbjct: 182 VLQNRSITVPIIHSSSEDNAYAQLLHLTSLSGILRNI 292
>BG653065 similar to GP|11139548|gb protein kinase AKINbetagamma-2 {Zea
mays}, partial (20%)
Length = 416
Score = 92.4 bits (228), Expect(2) = 9e-23
Identities = 52/88 (59%), Positives = 56/88 (63%)
Frame = +1
Query: 367 NIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQDAFN 426
N+ AQVSSIPIVD++DSLLDIYCRSDITALAK+RAYTHINLDEM
Sbjct: 55 NLLVQAQVSSIPIVDDNDSLLDIYCRSDITALAKNRAYTHINLDEM-------------- 192
Query: 427 PNESRSQRCQMCLRTDSLHKVMERLANP 454
TD LHKVMERLANP
Sbjct: 193 --------------TDPLHKVMERLANP 234
Score = 32.7 bits (73), Expect(2) = 9e-23
Identities = 19/23 (82%), Positives = 19/23 (82%)
Frame = +2
Query: 343 IAGL*QH*DQMLLLLQP*IY*FK 365
I GL*Q *DQ LLL QP*IY*FK
Sbjct: 2 IGGL*QC*DQPLLLRQP*IY*FK 70
>CA819632
Length = 353
Score = 68.9 bits (167), Expect = 4e-12
Identities = 32/48 (66%), Positives = 38/48 (78%)
Frame = +2
Query: 258 RRFIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASL 305
RRFIHAGP DNLKD+A+KILQ +STVPIIHSSS D S P +++ L
Sbjct: 2 RRFIHAGPYDNLKDIAMKILQKEVSTVPIIHSSSEDASAPNIVNFKKL 145
>TC211347 similar to UP|Q84PE0 (Q84PE0) AKIN betagamma, partial (16%)
Length = 666
Score = 60.5 bits (145), Expect = 2e-09
Identities = 29/46 (63%), Positives = 37/46 (80%), Gaps = 1/46 (2%)
Frame = +3
Query: 410 DEMTVHQALQLSQDAFNPNES-RSQRCQMCLRTDSLHKVMERLANP 454
DE+++HQAL L QDA +P+ QRC MCLR++SL+KVMERLANP
Sbjct: 3 DEISIHQALLLGQDATSPSGIYNGQRCHMCLRSESLYKVMERLANP 140
>TC218790 similar to UP|Q7XYX7 (Q7XYX7) AKIN beta2, partial (89%)
Length = 1533
Score = 52.4 bits (124), Expect = 4e-07
Identities = 31/101 (30%), Positives = 47/101 (45%)
Frame = +1
Query: 23 IPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEW 82
IP W Y G+ V + GS+ W + P++ F ++ L G +Q++F VDG+W
Sbjct: 625 IPTMITWSYDGKEVAVEGSWDNWKTRM---PLQRSGKDFTIMKVLPSGVYQFRFIVDGQW 795
Query: 83 RHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDIVSGSNMD 123
R+ P D G VL D +P DI S S+ +
Sbjct: 796 RYAPDLPWAQDDSGNAYNVLDLQD----YVPEDIGSISSFE 906
>TC206653 similar to UP|Q7XYX8 (Q7XYX8) AKIN beta1, partial (97%)
Length = 1071
Score = 52.0 bits (123), Expect = 6e-07
Identities = 32/96 (33%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Frame = +1
Query: 23 IPVRFVWPYGGRTVYLSGSFTRWS--ELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDG 80
IPV W YGG V + GS+ W+ + LQ S + ++ L PG + Y+F VDG
Sbjct: 520 IPVMITWNYGGNNVAVEGSWDNWTSRKALQRSGKD-----HSILIVLPPGIYHYRFIVDG 684
Query: 81 EWRHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDI 116
E R P++ + G V +L D +VP P +
Sbjct: 685 EERFTPQLPNVPDEMGHVCNLLDVND-YVPENPDGV 789
>BU550514 weakly similar to GP|8778877|gb| T7N9.13 {Arabidopsis thaliana},
partial (19%)
Length = 526
Score = 51.6 bits (122), Expect = 7e-07
Identities = 29/79 (36%), Positives = 41/79 (51%)
Frame = -1
Query: 28 VWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWRHDEH 87
VWP V L+GSF WS +M + +F + L PG ++ KF VDGEW+ D
Sbjct: 313 VWPNSASEVLLTGSFDGWSTKRKMERLSS--GIFSLNLQLYPGRYEMKFIVDGEWKIDPL 140
Query: 88 TPHITGDYGIVNTVLLATD 106
P +T + G N +L+ D
Sbjct: 139 RPVVTSN-GYENNLLIIYD 86
>TC206652 similar to UP|Q7XYX8 (Q7XYX8) AKIN beta1, partial (96%)
Length = 1242
Score = 49.7 bits (117), Expect = 3e-06
Identities = 30/94 (31%), Positives = 44/94 (45%)
Frame = +3
Query: 23 IPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEW 82
IPV W YGG V + GS+ W+ + G F ++ L PG + Y+F DGE
Sbjct: 573 IPVMITWNYGGNNVAVEGSWDNWTSRKALQRA-GKDHSFLIV--LPPGIYHYRFIADGEE 743
Query: 83 RHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDI 116
R P++ + G V +L D +VP P +
Sbjct: 744 RFIPELPNVADEMGHVCNLLDVND-YVPENPDGV 842
>BG651924 weakly similar to GP|8778877|gb| T7N9.13 {Arabidopsis thaliana},
partial (21%)
Length = 384
Score = 49.7 bits (117), Expect = 3e-06
Identities = 28/79 (35%), Positives = 41/79 (51%)
Frame = +3
Query: 28 VWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWRHDEH 87
VWP V L+GSF WS +M + +F + L PG ++ KF V+GEW+ D
Sbjct: 138 VWPNSASEVLLTGSFDGWSTKRKMERLSS--GIFSLNLQLYPGRYEMKFIVNGEWKIDPL 311
Query: 88 TPHITGDYGIVNTVLLATD 106
P +T + G N +L+ D
Sbjct: 312 RPVVTSN-GYENNLLIIYD 365
>CD415370
Length = 649
Score = 45.4 bits (106), Expect = 5e-05
Identities = 29/81 (35%), Positives = 40/81 (48%), Gaps = 2/81 (2%)
Frame = -3
Query: 25 VRFVWP-YGGRTVYLSGSFT-RWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEW 82
V FVW + G V L G FT W E L+ G +V L G + YKF V+G+W
Sbjct: 590 VTFVWNGHEGEDVTLVGDFTGNWKEPLKAKHQGGSRHEVEV--KLPQGKYYYKFIVNGQW 417
Query: 83 RHDEHTPHITGDYGIVNTVLL 103
+H +P D G VN +++
Sbjct: 416 KHSTASPAERDDKGNVNNIIV 354
>CA938353 similar to GP|15010688|gb AT5g39790/MKM21_80 {Arabidopsis
thaliana}, partial (40%)
Length = 406
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/83 (33%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Frame = +1
Query: 25 VRFVWPYGGRTVYLSGSFTRWSELLQMSP-VEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
V W +V + G+F WS+ +SP G T F L PG ++ KF VDGEW
Sbjct: 91 VSVFWVGMAESVQVMGTFDGWSQGEHLSPEYTGSYTRFSTTLLLRPGRYEIKFLVDGEWH 270
Query: 84 HDEHTPHITGDYGIVNTVLLATD 106
P I G+ G+ LL +
Sbjct: 271 LSPEFP-IIGE-GLTKNNLLVVE 333
>TC222796 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partial (23%)
Length = 405
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/60 (36%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Frame = +1
Query: 25 VRFVWPYGGRTVYLSGSFTRWSELLQMSP-VEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
V W +V + G+F WS+ +SP G T F L PG ++ KF VDGEW+
Sbjct: 1 VSVFWVGMAESVQVMGTFDGWSQGEHLSPEYTGSYTRFSTTLLLRPGRYEIKFLVDGEWK 180
>BU548295 similar to GP|15010688|gb| AT5g39790/MKM21_80 {Arabidopsis
thaliana}, partial (25%)
Length = 436
Score = 36.6 bits (83), Expect = 0.024
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 1/74 (1%)
Frame = -3
Query: 34 RTVYLSGSFTRWSELLQMSP-VEGCPTVFQVIHNLAPGYHQYKFFVDGEWRHDEHTPHIT 92
++V + G+ WS+ +SP G T F L PG ++ KF VDGEW P I
Sbjct: 350 QSVQVMGTXDGWSQGEHLSPEYTGSYTRFSTTLLLRPGRYEIKFLVDGEWHLSPEFP-II 174
Query: 93 GDYGIVNTVLLATD 106
G+ G+ LL +
Sbjct: 173 GE-GLTKNNLLVVE 135
>TC223721 similar to UP|Q9LFY0 (Q9LFY0) T7N9.13, partial (10%)
Length = 416
Score = 36.6 bits (83), Expect = 0.024
Identities = 18/47 (38%), Positives = 27/47 (57%)
Frame = +3
Query: 60 VFQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATD 106
+F + L PG ++ KF VDGEW+ D P +T + G N +L+ D
Sbjct: 246 IFSLNLQLYPGRYEMKFIVDGEWKIDPLRPVVTSN-GYENNLLIIYD 383
>CF806965
Length = 421
Score = 30.8 bits (68), Expect = 1.3
Identities = 19/57 (33%), Positives = 27/57 (47%)
Frame = +2
Query: 67 LAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATDPFVPVLPPDIVSGSNMD 123
L G +QY+F VDG R+ +P D G +L D +P DI S S+ +
Sbjct: 11 LPSGVYQYRFIVDGRMRYTPDSPWAQDDAGDAYNILDLQD----YVPEDIGSISSFE 169
>TC223959 similar to UP|Q9LFY0 (Q9LFY0) T7N9.13, partial (5%)
Length = 922
Score = 30.8 bits (68), Expect = 1.3
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = -1
Query: 73 QYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATD 106
Q KF VDGEW+ D P +T + G N +L+ D
Sbjct: 832 QMKFIVDGEWKIDPLRPVVTSN-GYENNLLIVHD 734
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,219,241
Number of Sequences: 63676
Number of extensions: 316931
Number of successful extensions: 1822
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1799
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1807
length of query: 454
length of database: 12,639,632
effective HSP length: 100
effective length of query: 354
effective length of database: 6,272,032
effective search space: 2220299328
effective search space used: 2220299328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146552.9


