

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.7 + phase: 0 /pseudo
(720 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
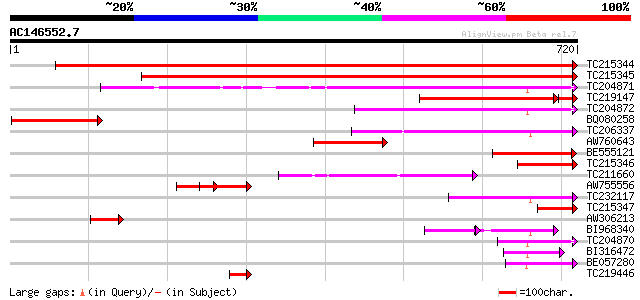
Score E
Sequences producing significant alignments: (bits) Value
TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 1256 0.0
TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 1045 0.0
TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%) 334 7e-92
TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11... 290 2e-85
TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%) 211 7e-55
BQ080258 204 9e-53
TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), par... 190 2e-48
AW760643 171 8e-43
BE555121 similar to GP|22335691|db cullin-like protein1 {Pisum s... 162 4e-40
TC215346 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 145 5e-35
TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), par... 131 9e-31
AW755556 homologue to GP|22335691|dbj cullin-like protein1 {Pisu... 110 2e-24
TC232117 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), par... 96 7e-20
TC215347 homologue to UP|Q711G6 (Q711G6) Cullin 1C (Fragment), p... 91 1e-18
AW306213 GP|22335691|db cullin-like protein1 {Pisum sativum}, pa... 90 3e-18
BI968340 44 3e-11
TC204870 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (17%) 66 6e-11
BI316472 57 3e-08
BE057280 similar to GP|9295728|gb| T24P13.25 {Arabidopsis thalia... 50 3e-06
TC219446 similar to UP|Q711G7 (Q711G7) Cullin 1B, partial (4%) 45 1e-04
>TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (92%)
Length = 2465
Score = 1256 bits (3251), Expect = 0.0
Identities = 628/662 (94%), Positives = 645/662 (96%)
Frame = +1
Query: 59 YSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDR 118
YSQ LYDKYKE+FEEYIV TVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDR
Sbjct: 1 YSQQLYDKYKESFEEYIVLTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDR 180
Query: 119 YFIARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIF 178
YFIARRSLPPLNEVGL CFRDL+YKEL+GK+RDA+ISLIDQEREGEQIDRALLKNVLDIF
Sbjct: 181 YFIARRSLPPLNEVGLTCFRDLIYKELNGKVRDAVISLIDQEREGEQIDRALLKNVLDIF 360
Query: 179 VEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAH 238
VEIGMG+MDHYENDFEA MLKDTSAYYSRKASNWILEDSCPDYMLKAEECL REKDRVAH
Sbjct: 361 VEIGMGQMDHYENDFEAAMLKDTSAYYSRKASNWILEDSCPDYMLKAEECL*REKDRVAH 540
Query: 239 YLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGL 298
YLHSSSEPKLLEKVQ+ELLSVYA+QLLEKEHSGCHALLRDDK EDLSRMFRLFSKIPRGL
Sbjct: 541 YLHSSSEPKLLEKVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLSRMFRLFSKIPRGL 720
Query: 299 DPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVN 358
DPVS+IFKQHVTTEGMALVK AEDAASNKKAEK+DIVG QEQVFVRKVIELHDKYLAYVN
Sbjct: 721 DPVSNIFKQHVTTEGMALVKQAEDAASNKKAEKKDIVGLQEQVFVRKVIELHDKYLAYVN 900
Query: 359 SCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLE 418
CFQNHTLFHKALKEAFEVFCNKGVAG+SSAELLA+FCDNILKKGGSEKLSDEAIEETLE
Sbjct: 901 DCFQNHTLFHKALKEAFEVFCNKGVAGSSSAELLASFCDNILKKGGSEKLSDEAIEETLE 1080
Query: 419 KVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGM 478
KVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGM
Sbjct: 1081KVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGM 1260
Query: 479 VTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEV 538
VTDLTLAKENQTSFEEYLSN PNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEM++CVEV
Sbjct: 1261VTDLTLAKENQTSFEEYLSNNPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMIRCVEV 1440
Query: 539 FKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEI 598
FKEFY TKTKHRKLTWIYSLGTCNISGKFDPKTVEL+VTTYQASALLLFNSSDRLSYSEI
Sbjct: 1441FKEFYQTKTKHRKLTWIYSLGTCNISGKFDPKTVELIVTTYQASALLLFNSSDRLSYSEI 1620
Query: 599 MTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPP 658
MTQLNL D+DVIRLLHSLSCAKYKIL KEPNTKTI TDYFEFN KFTDKMRRIKIPLPP
Sbjct: 1621MTQLNLSDDDVIRLLHSLSCAKYKILNKEPNTKTISSTDYFEFNYKFTDKMRRIKIPLPP 1800
Query: 659 VDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KR 718
VDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLV+ECVEQLGRMFKPDVKAI KR
Sbjct: 1801VDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVVECVEQLGRMFKPDVKAIKKR 1980
Query: 719 IE 720
IE
Sbjct: 1981IE 1986
>TC215345 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (77%)
Length = 2106
Score = 1045 bits (2702), Expect = 0.0
Identities = 525/553 (94%), Positives = 537/553 (96%)
Frame = +1
Query: 168 RALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEE 227
RALLKNVLDIFVEIGMG+MDHYENDFEA MLKDTSAYYSRKASNWILEDSCPDYMLKAEE
Sbjct: 1 RALLKNVLDIFVEIGMGQMDHYENDFEAAMLKDTSAYYSRKASNWILEDSCPDYMLKAEE 180
Query: 228 CLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRM 287
CL+REKDRVAHYLHSSSEPKLLEKVQ+ELLSVYA+QLLEKEHSGCHALLRDDK EDLSRM
Sbjct: 181 CLKREKDRVAHYLHSSSEPKLLEKVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLSRM 360
Query: 288 FRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVI 347
FRLFSKIPRGLDPVSSIFKQHVT EGMALVK AEDA S KKAEK+DIVG QEQVFVRKVI
Sbjct: 361 FRLFSKIPRGLDPVSSIFKQHVTAEGMALVKLAEDAVSTKKAEKKDIVGLQEQVFVRKVI 540
Query: 348 ELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEK 407
ELHDKYLAYVN CFQNHTLFHKALKEAFE+FCNKGVAG+SSAELLATFCDNILKKGGSEK
Sbjct: 541 ELHDKYLAYVNDCFQNHTLFHKALKEAFEIFCNKGVAGSSSAELLATFCDNILKKGGSEK 720
Query: 408 LSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQC 467
LSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQC
Sbjct: 721 LSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQC 900
Query: 468 GGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLN 527
GGQFTSKMEGMVTDLTLAKENQTSFEEYL+N PNADPGIDLTVTVLTTGFWPSYKSFDLN
Sbjct: 901 GGQFTSKMEGMVTDLTLAKENQTSFEEYLTNNPNADPGIDLTVTVLTTGFWPSYKSFDLN 1080
Query: 528 LPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLF 587
LPAEMV+CVEVFKEFY TKTKHRKLTWIYSLGTCNISGKFDPKTVEL+VTTYQASALLLF
Sbjct: 1081LPAEMVRCVEVFKEFYQTKTKHRKLTWIYSLGTCNISGKFDPKTVELIVTTYQASALLLF 1260
Query: 588 NSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTD 647
NSSDRLSYSEIMTQLNL D+DVIRLLHSLSCAKYKIL KEPNTKTI TDYFEFN+KFTD
Sbjct: 1261NSSDRLSYSEIMTQLNLSDDDVIRLLHSLSCAKYKILNKEPNTKTISSTDYFEFNSKFTD 1440
Query: 648 KMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRM 707
KMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVL YQQLVMECVEQLGRM
Sbjct: 1441KMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLNYQQLVMECVEQLGRM 1620
Query: 708 FKPDVKAI*KRIE 720
FKPDVKAI KRIE
Sbjct: 1621FKPDVKAIKKRIE 1659
>TC204871 similar to UP|Q84LL5 (Q84LL5) Cullin 4, partial (81%)
Length = 2087
Score = 334 bits (857), Expect = 7e-92
Identities = 209/613 (34%), Positives = 334/613 (54%), Gaps = 8/613 (1%)
Frame = +1
Query: 116 LDRYFIARRS-LPPLNEVGLACFRD--LVYKELHGKMRDAIISLIDQEREGEQIDRALLK 172
LDR ++ + + + L ++GL FR + E+ K ++ +I+ ER+GE +DR LL
Sbjct: 1 LDRTYVKQTANVRSLWDMGLQLFRKHLSLSPEVEHKTVTGLLRMIESERKGEAVDRTLLN 180
Query: 173 NVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRRE 232
++L +F +G+ Y FE L+ TS +Y+ + ++ + PDY+ E L+ E
Sbjct: 181 HLLKMFTALGI-----YAESFEKPFLECTSEFYAAEGVKYMQQSDVPDYLKHVEIRLQEE 345
Query: 233 KDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFS 292
+R YL +S+ L+ + +LL + +L+K G L+ ++ EDL RM+ LFS
Sbjct: 346 HERCLIYLDASTRKPLIATAEKQLLERHIPAILDK---GFAMLMDGNRIEDLQRMYSLFS 516
Query: 293 KIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDK 352
++ L+ + ++ G +V E +++ V ++E
Sbjct: 517 RV-NALESLRQAISSYIRRTGQGIVLDEE----------------KDKDMVSSLLEFKAS 645
Query: 353 YLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEA 412
F + F +K++FE N + N AEL+A F D L+ G++ S+E
Sbjct: 646 LDTTWEESFSKNEAFCNTIKDSFEHLIN--LRQNRPAELIAKFLDEKLR-AGNKGTSEEE 816
Query: 413 IEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFT 472
+E TL+KV+ L +I KD+F FY+K LA+RLL KSA+ D E+S+++KLK +CG QFT
Sbjct: 817 LEGTLDKVLVLFRFIQGKDVFEAFYKKDLAKRLLLGKSASIDAEKSMISKLKTECGSQFT 996
Query: 473 SKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEM 532
+K+EGM D+ L+KE SF++ GI+++V VLTTG+WP+Y D+ LP E+
Sbjct: 997 NKLEGMFKDIELSKEINESFKQSSQARTKLPSGIEMSVHVLTTGYWPTYPPMDVRLPHEL 1176
Query: 533 VKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDR 592
++FKEFY +K R+L W SLG C + +F EL V+ +Q L+LFN +++
Sbjct: 1177NVYQDIFKEFYLSKYSGRRLMWQNSLGHCVLKAEFPKGKKELAVSLFQTVVLMLFNDAEK 1356
Query: 593 LSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRI 652
LS+ +I + D+++ R L SL+C K ++L K P + + D F FN FT + RI
Sbjct: 1357LSFQDIKDSTGIEDKELRRTLQSLACGKVRVLQKLPKGRDVEDDDSFVFNEGFTAPLYRI 1536
Query: 653 KIPL----PPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMF 708
K+ V+E E V +DR+Y +DA+IVRIMK+RKVL + L+ E +QL
Sbjct: 1537KVNAIQLKETVEENTSTTERVFQDRQYQVDAAIVRIMKTRKVLSHTLLITELFQQLKFPI 1716
Query: 709 KP-DVKAI*KRIE 720
KP D+K KRIE
Sbjct: 1717KPADLK---KRIE 1746
>TC219147 similar to PIR|T01092|T01092 cullin-like protein T10P11.14.1 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(33%)
Length = 1237
Score = 290 bits (743), Expect(2) = 2e-85
Identities = 141/177 (79%), Positives = 160/177 (89%)
Frame = +3
Query: 521 YKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQ 580
YKSFDLNLP+EM++C+EVFK FY T+TKHRKLTWIYSLGTC+++GKF+ K +EL+V TY
Sbjct: 3 YKSFDLNLPSEMIRCLEVFKGFYETRTKHRKLTWIYSLGTCHVTGKFETKNIELIVPTYP 182
Query: 581 ASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFE 640
A+ALLLFN++DRLSYSEIMTQLNL EDV RLLHSLS AKYKILIKEPN K I +D FE
Sbjct: 183 AAALLLFNNADRLSYSEIMTQLNLGHEDVARLLHSLSSAKYKILIKEPNNKVISQSDIFE 362
Query: 641 FNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLV 697
FN KFTDKMRRIKIPLPP DE+KKVIEDVDKDRRYAIDA+IVRIMKSRK+LG+QQLV
Sbjct: 363 FNYKFTDKMRRIKIPLPPADERKKVIEDVDKDRRYAIDAAIVRIMKSRKILGHQQLV 533
Score = 45.1 bits (105), Expect(2) = 2e-85
Identities = 20/23 (86%), Positives = 22/23 (94%)
Frame = +1
Query: 698 MECVEQLGRMFKPDVKAI*KRIE 720
+ECVEQLGRMFKPD+KAI KRIE
Sbjct: 535 LECVEQLGRMFKPDIKAIKKRIE 603
>TC204872 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (42%)
Length = 1224
Score = 211 bits (538), Expect = 7e-55
Identities = 122/288 (42%), Positives = 172/288 (59%), Gaps = 5/288 (1%)
Frame = +2
Query: 438 RKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLS 497
RK L RRLL KSA+ D E+S+++KLK +CG QFT+K+EGM D+ L+KE SF++
Sbjct: 17 RKILLRRLLLGKSASIDAEKSMISKLKTECGSQFTNKLEGMFKDIELSKEINDSFKQSSQ 196
Query: 498 NTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYS 557
GI+++V VLTTG WP+Y D+ LP E+ ++FKEFY +K R+L W S
Sbjct: 197 ARSKLASGIEMSVHVLTTGHWPTYPPMDVRLPHELNVYQDIFKEFYLSKYSGRRLMWQNS 376
Query: 558 LGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLS 617
LG C + +F EL V+ +Q L+LFN +++LS +I + D+++ R L SL+
Sbjct: 377 LGHCVLKAEFPKGRKELAVSLFQTVVLMLFNDAEKLSLQDIKDATGIEDKELRRTLQSLA 556
Query: 618 CAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPL----PPVDEKKKVIEDVDKDR 673
C K ++L K P + + D F FN FT + RIK+ V+E E V DR
Sbjct: 557 CGKVRVLQKMPKGRDVEDDDLFVFNDGFTAPLYRIKVNAIQLKETVEENTSTTERVFHDR 736
Query: 674 RYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP-DVKAI*KRIE 720
+Y IDA+IVRIMK+RKVL + L+ E +QL KP D+K KRIE
Sbjct: 737 QYQIDAAIVRIMKTRKVLSHTLLITELFQQLKFPIKPADLK---KRIE 871
>BQ080258
Length = 434
Score = 204 bits (520), Expect = 9e-53
Identities = 93/115 (80%), Positives = 102/115 (87%)
Frame = +3
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERK ID +QGWD+M +GI KLK ILEG PE FS E+YMMLYTTIYNMCTQKPPND+SQ
Sbjct: 90 ERKIIDFDQGWDYMQKGITKLKKILEGAPETPFSSEEYMMLYTTIYNMCTQKPPNDFSQQ 269
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLD 117
LYDKYK+AF+EYI TVLPSLREKHDEFMLRELV+RW NHK+MVRWLSRFFHYLD
Sbjct: 270 LYDKYKDAFDEYIKITVLPSLREKHDEFMLRELVQRWLNHKVMVRWLSRFFHYLD 434
>TC206337 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), partial (91%)
Length = 1135
Score = 190 bits (483), Expect = 2e-48
Identities = 117/293 (39%), Positives = 169/293 (56%), Gaps = 7/293 (2%)
Frame = +3
Query: 435 EFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEE 494
++Y++ LA+RLL K+ +DD ERS++ KLK +CG QFTSK+EGM TD+ + + F
Sbjct: 3 KYYKQHLAKRLLSGKTISDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSHDTMQGFYA 182
Query: 495 YLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTW 554
L G L+V VLTTG WP+ S NLP E++ + F+ +Y R+L+W
Sbjct: 183 ILGT--ELGDGPLLSVQVLTTGSWPTQPSPPCNLPVEILGVCDKFRTYYLGTHNGRRLSW 356
Query: 555 IYSLGTCNISGKFDP-KTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLL 613
++GT ++ F + EL V+TYQ L+LFNS++RL+ EI + D+ R L
Sbjct: 357 QTNMGTADLKATFGKGQKHELNVSTYQMCVLMLFNSAERLTCKEIEQATAIPMSDLRRCL 536
Query: 614 HSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD-----EKKKVIE 667
SL+C K K +L KEP +K I D F FN KFT K ++KI E + +
Sbjct: 537 QSLACVKGKNVLRKEPMSKDIAEDDAFFFNDKFTSKFFKVKIGTVVAQRESEPENLETRQ 716
Query: 668 DVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
V++DR+ I+A+IVRIMKSR+ L + +V E +QL F P+ I KRIE
Sbjct: 717 RVEEDRKPQIEAAIVRIMKSRRTLDHNNIVAEVTKQLQSRFLPNPVVIKKRIE 875
>AW760643
Length = 286
Score = 171 bits (434), Expect = 8e-43
Identities = 86/93 (92%), Positives = 90/93 (96%)
Frame = +1
Query: 387 SSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLL 446
SSAELL+TFCDNILKKGGSEKLSDEAIE+TLEKVVKLLAYISDKDLFAEFYR KLARRLL
Sbjct: 4 SSAELLSTFCDNILKKGGSEKLSDEAIEDTLEKVVKLLAYISDKDLFAEFYRNKLARRLL 183
Query: 447 FDKSANDDHERSILTKLKQQCGGQFTSKMEGMV 479
D+SANDDHE+ ILTKLKQQCGGQFTSKMEGMV
Sbjct: 184 CDRSANDDHEKCILTKLKQQCGGQFTSKMEGMV 282
>BE555121 similar to GP|22335691|db cullin-like protein1 {Pisum sativum},
partial (17%)
Length = 388
Score = 162 bits (411), Expect = 4e-40
Identities = 82/106 (77%), Positives = 90/106 (84%)
Frame = +2
Query: 614 HSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKVIEDVDKDR 673
HSLSCAK KIL KEPNTKT+ TDY E+N KFTDKMRRIKIP PPVDEKKK+IED D R
Sbjct: 2 HSLSCAKNKILNKEPNTKTMSYTDYSEYNPKFTDKMRRIKIPFPPVDEKKKLIEDADNYR 181
Query: 674 RYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRI 719
RYA+DA+IVR++ SRKVL +QQLVMECVEQLG MFKPDV AI K I
Sbjct: 182 RYAVDATIVRLINSRKVLSHQQLVMECVEQLGHMFKPDVIAIKKLI 319
>TC215346 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (13%)
Length = 1050
Score = 145 bits (367), Expect = 5e-35
Identities = 74/76 (97%), Positives = 74/76 (97%)
Frame = +1
Query: 645 FTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQL 704
FTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVL YQQLVMECVEQL
Sbjct: 1 FTDKMRRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLSYQQLVMECVEQL 180
Query: 705 GRMFKPDVKAI*KRIE 720
GRMFKPDVKAI KRIE
Sbjct: 181 GRMFKPDVKAIKKRIE 228
>TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), partial (29%)
Length = 747
Score = 131 bits (330), Expect = 9e-31
Identities = 81/254 (31%), Positives = 138/254 (53%), Gaps = 1/254 (0%)
Frame = +1
Query: 342 FVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILK 401
FV+++++L DKY + F N F AL +FE F N S E ++ F D+ L+
Sbjct: 4 FVQRLLDLKDKYDRVITMSFNNDKTFQNALNSSFEYFINLNAR---SPEFISLFVDDKLR 174
Query: 402 KGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILT 461
+G + + +E +E L KV+ L Y+ +KD+F ++Y++ LA+RLL K+ + D ERS++
Sbjct: 175 RG-LKGVGEEDVEIVLNKVMMLFRYLQEKDVFEKYYKQHLAKRLLSRKTISYDAERSLIV 351
Query: 462 KLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSY 521
KL+ QCG QFTSK++GM TD+T + + Y + G L+V VLTTG WP+
Sbjct: 352 KLETQCGYQFTSKLQGMFTDMTTSHDTMQGL--YANLGSELGDGPMLSVQVLTTGSWPTQ 525
Query: 522 KSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKF-DPKTVELVVTTYQ 580
+ NL +++ + + +Y R+L + ++ T + + + E Y
Sbjct: 526 PTPPCNLTV*ILRVSDKSRTYYLDSHNGRRLYSLTNIETA*VKTTLCNSQAYEFH*FVYG 705
Query: 581 ASALLLFNSSDRLS 594
S L+L+ S +R++
Sbjct: 706 MSVLVLYCSIERVT 747
>AW755556 homologue to GP|22335691|dbj cullin-like protein1 {Pisum sativum},
partial (12%)
Length = 442
Score = 110 bits (275), Expect = 2e-24
Identities = 57/66 (86%), Positives = 60/66 (90%)
Frame = +2
Query: 242 SSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPV 301
+SS L KVQ+ELLSVYA+QLLEKEHSGCHALLRDDK EDLSRMFRLFSKIPRGLDPV
Sbjct: 245 NSSVCFLF*KVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLSRMFRLFSKIPRGLDPV 424
Query: 302 SSIFKQ 307
SSIFKQ
Sbjct: 425 SSIFKQ 442
Score = 85.9 bits (211), Expect = 6e-17
Identities = 40/54 (74%), Positives = 47/54 (86%)
Frame = +1
Query: 212 WILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLL 265
WILEDSCPDYMLKAEECL+REKDRVAHYLHSSSEPKLLE + ++++S +L
Sbjct: 1 WILEDSCPDYMLKAEECLKREKDRVAHYLHSSSEPKLLEVWFALISAIWSSTVL 162
>TC232117 similar to UP|Q7Y1Y8 (Q7Y1Y8) Cullin 3a (Fragment), partial (57%)
Length = 832
Score = 95.5 bits (236), Expect = 7e-20
Identities = 64/170 (37%), Positives = 94/170 (54%), Gaps = 7/170 (4%)
Frame = +1
Query: 558 LGTCNISGKFDP-KTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSL 616
+GT ++ F + EL V+TYQ L+LFN++DR Y EI + D+ R L SL
Sbjct: 1 MGTADLKATFGKGQKHELNVSTYQMCVLMLFNNADRXXYKEIEQATEIPASDLKRCLQSL 180
Query: 617 SCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVD 670
+ K + +L KEP K I D F N KF+ K+ ++KI EK++ + V+
Sbjct: 181 ALVKGRNVLRKEPMGKDIGDDDAFYVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVE 360
Query: 671 KDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
+DR+ I+A+IVRIMKSRK L + L+ E +QL F + + KRIE
Sbjct: 361 EDRKPQIEAAIVRIMKSRKQLDHNNLIAEVTKQLQSRFLANPTEVKKRIE 510
>TC215347 homologue to UP|Q711G6 (Q711G6) Cullin 1C (Fragment), partial (15%)
Length = 668
Score = 91.3 bits (225), Expect = 1e-18
Identities = 47/50 (94%), Positives = 47/50 (94%)
Frame = +1
Query: 671 KDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
K RYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI KRIE
Sbjct: 37 KGGRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAIKKRIE 186
>AW306213 GP|22335691|db cullin-like protein1 {Pisum sativum}, partial (5%)
Length = 127
Score = 90.1 bits (222), Expect = 3e-18
Identities = 41/42 (97%), Positives = 41/42 (97%)
Frame = +1
Query: 103 KIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLACFRDLVYKE 144
KIMVRWLSRFFHYLDRYFIARRSLPPLNEVGL CFRDLVYKE
Sbjct: 1 KIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLTCFRDLVYKE 126
>BI968340
Length = 621
Score = 44.3 bits (103), Expect(2) = 3e-11
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Frame = -2
Query: 527 NLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDP-KTVELVVTTYQASALL 585
NLP EM+ + F +Y R+L+W ++G ++ F + EL V+T Q L+
Sbjct: 533 NLPVEMLGVCDKFGTYYLGSHNGRRLSWQGNMGGGDLKAAFGKGQKHELNVSTCQMCVLM 354
Query: 586 LFNSSDRLSYSEI 598
LFNS +RL+ EI
Sbjct: 353 LFNSXERLTCKEI 315
Score = 42.7 bits (99), Expect(2) = 3e-11
Identities = 34/113 (30%), Positives = 52/113 (45%), Gaps = 6/113 (5%)
Frame = -1
Query: 591 DRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKM 649
DR YS + +L ++C K K +L KEP +K I D FN KF K+
Sbjct: 318 DRAGYSNTHVRFE-------EVLQCVACVKGKNVLRKEPMSKDIXEDDALFFNGKFASKL 160
Query: 650 RRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLV 697
+KI E + + V++DR+ I +SIV +KS + L + L+
Sbjct: 159 FXVKIGTVVAQRESEPENLETRQRVEEDRKPQIXSSIVXXIKSXRTLYHNILL 1
>TC204870 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (17%)
Length = 621
Score = 65.9 bits (159), Expect = 6e-11
Identities = 44/106 (41%), Positives = 60/106 (56%), Gaps = 5/106 (4%)
Frame = +2
Query: 620 KYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPL----PPVDEKKKVIEDVDKDRRY 675
K ++L K P + + D F FN FT + RIK+ V+E E V +DR+Y
Sbjct: 5 KVRVLPKLPKGRDVEDDDSFVFNEGFTAPLYRIKVNAIQLKETVEENTSTTERVFQDRQY 184
Query: 676 AIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP-DVKAI*KRIE 720
+DA+IVRIMK+RKVL + L+ E +QL KP D+K KRIE
Sbjct: 185 QVDAAIVRIMKTRKVLSHTLLITELFQQLKFPIKPADLK---KRIE 313
>BI316472
Length = 415
Score = 57.0 bits (136), Expect = 3e-08
Identities = 34/81 (41%), Positives = 45/81 (54%), Gaps = 4/81 (4%)
Frame = +1
Query: 628 PNTKTILPTDYFEFNAKFTDKMRRIKIPL----PPVDEKKKVIEDVDKDRRYAIDASIVR 683
P + + D F FN FT + RIK+ V+E E V DR+Y IDA+IVR
Sbjct: 163 PKGRDVEDDDLFVFNDGFTAPLYRIKVNAIQLKETVEENTSTTERVFHDRQYQIDAAIVR 342
Query: 684 IMKSRKVLGYQQLVMECVEQL 704
IMK+RKVL + L+ E +QL
Sbjct: 343 IMKTRKVLSHTLLITELFQQL 405
>BE057280 similar to GP|9295728|gb| T24P13.25 {Arabidopsis thaliana}, partial
(16%)
Length = 403
Score = 50.4 bits (119), Expect = 3e-06
Identities = 34/96 (35%), Positives = 49/96 (50%), Gaps = 5/96 (5%)
Frame = +3
Query: 630 TKTILPTDYFEFNAKFTDKMRRIKI-----PLPPVDEKKKVIEDVDKDRRYAIDASIVRI 684
+K I D F +N K T K+ +KI E ++ ++R I+A+IVRI
Sbjct: 15 SKDIAEDDAFFWNDKVTSKVCEVKIGSVGGEXESEPENVGTLQIXGEEREPQIEAAIVRI 194
Query: 685 MKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
MKSR+ L + +V E +QL F P+ I KRIE
Sbjct: 195 MKSRRTLDHNNIVAEVTKQLQSRFLPNPVVIKKRIE 302
>TC219446 similar to UP|Q711G7 (Q711G7) Cullin 1B, partial (4%)
Length = 955
Score = 45.1 bits (105), Expect = 1e-04
Identities = 19/28 (67%), Positives = 25/28 (88%)
Frame = +1
Query: 280 KCEDLSRMFRLFSKIPRGLDPVSSIFKQ 307
K EDLSRM+RL+ K P+GLDPV+++FKQ
Sbjct: 355 KVEDLSRMYRLYYKKPKGLDPVANVFKQ 438
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,251,291
Number of Sequences: 63676
Number of extensions: 386132
Number of successful extensions: 1929
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1911
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1920
length of query: 720
length of database: 12,639,632
effective HSP length: 104
effective length of query: 616
effective length of database: 6,017,328
effective search space: 3706674048
effective search space used: 3706674048
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146552.7


