

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.4 + phase: 0
(340 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
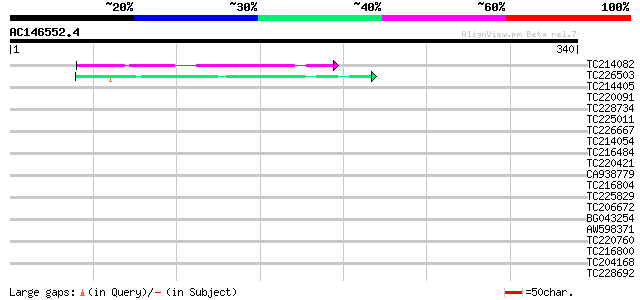
Score E
Sequences producing significant alignments: (bits) Value
TC214082 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, p... 44 8e-05
TC226503 similar to UP|Q6ILL4 (Q6ILL4) HDC09080, partial (6%) 41 7e-04
TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor T... 40 0.001
TC220091 similar to UP|Q94AE4 (Q94AE4) AT5g43400/MWF20_9 (Emb|CA... 40 0.002
TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%) 39 0.003
TC225011 similar to UP|Q09083 (Q09083) Hydroxyproline-rich glyco... 38 0.008
TC226667 similar to UP|Q9SE96 (Q9SE96) FH protein interacting pr... 37 0.010
TC214054 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, p... 36 0.022
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 35 0.064
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 35 0.064
CA938779 similar to GP|10177876|dbj contains similarity to prote... 35 0.064
TC216804 similar to UP|PFD5_ARATH (P57742) Probable prefoldin su... 34 0.083
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 34 0.11
TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyra... 34 0.11
BG043254 34 0.11
AW598371 weakly similar to GP|9280319|dbj| extensin protein-like... 33 0.14
TC220760 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (9%) 33 0.14
TC216800 weakly similar to UP|Q9LRQ7 (Q9LRQ7) Anthocyanin 5-arom... 33 0.19
TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime ... 33 0.19
TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, pa... 32 0.41
>TC214082 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, partial (34%)
Length = 1032
Score = 44.3 bits (103), Expect = 8e-05
Identities = 33/157 (21%), Positives = 64/157 (40%)
Frame = +1
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHN 100
PT + E + Q+ P P P T +P +T Q A+EV + T ST
Sbjct: 136 PTTVPENETTEVIKTQETTPEPVPATEAP--ATEQAAAEVPAQEEQQATEVPAPESTTE- 306
Query: 101 LEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHI 160
P + + P +ET T + PE+ + +E+V +T ++
Sbjct: 307 -----------APKEETTEAPTTETVETTTTEEAKPEEPKEVAVETEEVVTKETEEEKPA 453
Query: 161 PSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSP 197
+++ E+ ++ + +++P + T+TE +P
Sbjct: 454 AEEKSEEKTEE------VKEEAEEPKETTETETESAP 546
>TC226503 similar to UP|Q6ILL4 (Q6ILL4) HDC09080, partial (6%)
Length = 1597
Score = 41.2 bits (95), Expect = 7e-04
Identities = 43/187 (22%), Positives = 67/187 (34%), Gaps = 6/187 (3%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVSVQ-QPQ-----PNPEPTTNSPHNSTTQKASEVASDATTSETPQHQ 93
P T + NP+ + Q QP P+P SP S+ K S TTS + +
Sbjct: 223 PHTQISFATNPKSSAAQVQPSSSTQSPSPPVMVGSPTTSSMSK--NTGSPRTTSASTTNN 396
Query: 94 EYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQ 153
+ S +L P + S V + V +P+ T T S+ P +
Sbjct: 397 KISQSSSLSSQQAKNSSAVPARKSSPVGSRNV----PSILNVPQLTPSSSTGSKSQLPQK 564
Query: 154 TTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIPDHILESEYIEEQLI 213
Q IP + Q SP + P ++TT T P Y++ Q
Sbjct: 565 QQPQQQIPKQALQQAQLLFSSPYV------HPQSNSTTSTTTVP------SGYYLQHQQR 708
Query: 214 RLSDEIQ 220
R +++Q
Sbjct: 709 RGPEQMQ 729
>TC214405 homologue to UP|EFT1_SOYBN (Q43467) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 2050
Score = 40.4 bits (93), Expect = 0.001
Identities = 41/167 (24%), Positives = 54/167 (31%), Gaps = 7/167 (4%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVS-------VQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQH 92
PPT YP +P P S P P P P+ P S AS A T + P
Sbjct: 112 PPTPTNYPLSPPPSSTPPPSSAAPPPSPLPPPSRADPSPSAPPAASSSARSPT*TSAP-- 285
Query: 93 QEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPD 152
S + P P S T P ++ + + P + S
Sbjct: 286 ---SATWTTARPP*PPPSPWPLPPSATAPRRSTTRSTPPRRNAPAASPSTPPPSSTRPRT 456
Query: 153 QTTSDQHIPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIP 199
TT P+ TT + P P PS+S+ T P P P
Sbjct: 457 ATTPTWTAPATPTTSRT*SPAPP-----RWTAPSLSSPAPTAPCPKP 582
>TC220091 similar to UP|Q94AE4 (Q94AE4) AT5g43400/MWF20_9 (Emb|CAB86628.1),
partial (16%)
Length = 887
Score = 39.7 bits (91), Expect = 0.002
Identities = 38/161 (23%), Positives = 57/161 (34%), Gaps = 11/161 (6%)
Frame = +2
Query: 50 PEPVSVQQPQPNPEPTTNSP--------HNSTTQKASEVASDATTSETPQHQEYSTLHNL 101
P P P PNP PT NS H S + T P + S + +
Sbjct: 233 PHPNPSPNPNPNPNPTPNSHKNHSN*PFHRSNGGQIQHHVIPTTPKHDPNRKHVSHILHN 412
Query: 102 EKHLGGEMQPT-PTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHI 160
K + G + P P ++ P KT T P + S +A +
Sbjct: 413 GKSMSGLLLPRGPRHPTRDYPPKTRTCLGPSTPLPPSNSSATSAVSAALASPTVRASTPP 592
Query: 161 PSDQT--TEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIP 199
PS T T + P SP +L + + S+ ++T P+P
Sbjct: 593 PSGSTAVTPKPSLPTSPPSPNLDTLRISLRSSTSF*KDPMP 715
>TC228734 weakly similar to UP|Q99738 (Q99738) Pinin, partial (4%)
Length = 1156
Score = 38.9 bits (89), Expect = 0.003
Identities = 40/160 (25%), Positives = 61/160 (38%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
P T P P P P Q P P+P P + ST ASE + +T P S
Sbjct: 382 PSTSPP-PATPLPPLPQPPTPSPSPCRSPSPPSTPTTASEFTTPNSTPTPPTAASRSP-- 552
Query: 100 NLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQH 159
P PT+A+ T P T + + P ++ +++S P++ S
Sbjct: 553 ------SPPPSPPPTRATATPPSGPPTSTPPPSPSPPSRS---RSSSRTKPPEEFWST*K 705
Query: 160 IPSDQTTEQQQQPDSPTIIDLTSDQPSISNTTQTEPSPIP 199
+ + + TI+ T + S S T T PS +P
Sbjct: 706 STEESSGRSELGFPEFTILMSTVRRTSGSPATGTTPSDLP 825
>TC225011 similar to UP|Q09083 (Q09083) Hydroxyproline-rich glycoprotein
precursor, partial (20%)
Length = 964
Score = 37.7 bits (86), Expect = 0.008
Identities = 40/166 (24%), Positives = 60/166 (36%), Gaps = 10/166 (6%)
Frame = +1
Query: 39 LPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTL 98
+ PT +P P P P+P P+ P +STT + + +TTS P S+
Sbjct: 61 ISPTSSAPLPSPSPTPTPSPTPSPSPSLCLPLSSTTS*EAILLPFSTTSSPPPPLPPSS- 237
Query: 99 HNLEKHLGGEMQPTPTKASKTVPEKTVL----ETQTETQTIPEQTVQEQTASEQVAPDQT 154
P PT +S T + +L T + +P + P
Sbjct: 238 ------------PPPTPSSSTTS*EAILLSFSTTSSPPPPLPPSSPPPTPLPPSSPPPTP 381
Query: 155 TSDQHIPSDQTT--EQQQQPDSPTI----IDLTSDQPSISNTTQTE 194
+S +P +T E Q P +PT + T P S TT E
Sbjct: 382 SSSLRLPLSSSTP*EALQVPFTPTTSSPSLHSTPSLPLSSTTTLQE 519
>TC226667 similar to UP|Q9SE96 (Q9SE96) FH protein interacting protein FIP1
(At1g28200/F3H9_12), partial (83%)
Length = 1252
Score = 37.4 bits (85), Expect = 0.010
Identities = 26/88 (29%), Positives = 38/88 (42%)
Frame = +2
Query: 112 TPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQ 171
T T+A+ T PE+T TQT T E Q Q+ + D + P+D +
Sbjct: 128 TQTQATDTKPEETHTHTQTITTKPEESHSQSQSQPQPHTADYAPYPKLDPTDVAPPLNTE 307
Query: 172 PDSPTIIDLTSDQPSISNTTQTEPSPIP 199
+P D + P SN T P+P+P
Sbjct: 308 SRAPISEDAATTMPKDSNPYVT-PAPVP 388
>TC214054 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, partial (68%)
Length = 1306
Score = 36.2 bits (82), Expect = 0.022
Identities = 22/72 (30%), Positives = 32/72 (43%)
Frame = +1
Query: 114 TKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQTTEQQQQPD 173
T+ SKT V E Q E EQ A+E AP+ TT P ++TTE +
Sbjct: 151 TEVSKTQETTPVTEAPATEQPAAEVPATEQPAAEAPAPESTT---EAPKEETTEAPTETV 321
Query: 174 SPTIIDLTSDQP 185
T ++ ++P
Sbjct: 322 EKTTTEVAPEEP 357
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 34.7 bits (78), Expect = 0.064
Identities = 39/169 (23%), Positives = 63/169 (37%), Gaps = 21/169 (12%)
Frame = +1
Query: 44 PLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQE---YSTLHN 100
P P +P P + P P P+P +S ++T + S TT+ +PQ +L
Sbjct: 349 PTAPPSP-PQCTKPPAPPPDPPASSTSPTSTAEKSHSPPKTTTAHSPQRSSNP*KKSLTT 525
Query: 101 LEKHLGGEMQP----------TPTKASKTVPEKT------VLETQTETQTIPEQTVQEQT 144
+ + P P K S T+ + T L T+ P T
Sbjct: 526 SQ*SRSAKFSPLPQRKPLHPHPPNKTSPTLCQNTDARSSPTLSLLNPTRLTPSMTT-SMG 702
Query: 145 ASEQVAPDQTTSDQHIPSDQT--TEQQQQPDSPTIIDLTSDQPSISNTT 191
A AP T S + P+ +T + + + T+ TS +P + TT
Sbjct: 703 A*PSFAPSTTRSKRFSPNSKT*PNPVRSRFSNSTVFRFTSPKPLSNPTT 849
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 34.7 bits (78), Expect = 0.064
Identities = 26/112 (23%), Positives = 40/112 (35%)
Frame = +3
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
PP+ P P P P S P+ P T SP S AS ++++ S +P + T
Sbjct: 345 PPSSPSPPSAPSPSSTAPSPPSASPPT-SPSPSPPSPASTASANSPASGSPTSR---TSP 512
Query: 100 NLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAP 151
P P+ +S P + T P T+ ++P
Sbjct: 513 TSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISP 668
Score = 34.3 bits (77), Expect = 0.083
Identities = 33/162 (20%), Positives = 60/162 (36%), Gaps = 4/162 (2%)
Frame = +3
Query: 45 LYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKH 104
L+ +P P+ Q Q P+T+ PH + + T +P +T
Sbjct: 147 LHLHHPHPLWTQNKQQPLNPSTSPPHRTPAPSPPSTMPPSATPPSPS----ATSSPSASP 314
Query: 105 LGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQHIPSDQ 164
P+P+ S P + + T P + S +T+ + P+
Sbjct: 315 TAPPTSPSPSPPSSPSPPSA--PSPSSTAPSPPSASPPTSPSPSPPSPASTASANSPASG 488
Query: 165 TTEQQQQPDSPTIIDLTSDQPSISNTT----QTEPSPIPDHI 202
+ + P SP+ TS P+ S+++ ++PSP P I
Sbjct: 489 SPTSRTSPTSPSPTS-TSKPPAPSSSSPA*PSSKPSPSPTPI 611
>CA938779 similar to GP|10177876|dbj contains similarity to protein
kinase~gene_id:MIO24.10 {Arabidopsis thaliana}, partial
(6%)
Length = 434
Score = 34.7 bits (78), Expect = 0.064
Identities = 28/108 (25%), Positives = 44/108 (39%), Gaps = 3/108 (2%)
Frame = -3
Query: 40 PPTHPLYPDNPEPVSVQQ-PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYS-T 97
PP+H + + P Q+ PNP P +SP A +A +H E+S
Sbjct: 372 PPSHSAHYPSQTPARPQRRTPPNPSPAKSSP----PSPAKAIAPHPNHHPP*KHYEHSPP 205
Query: 98 LHNLEKHLGGEMQPTPTKASKT-VPEKTVLETQTETQTIPEQTVQEQT 144
+HN ++ P P+ +S + E Q +T P T Q Q+
Sbjct: 204 IHNHQQQQQQPPPPPPSSSSSLHYQHSHIPEPQFQTSESPSSTSQAQS 61
>TC216804 similar to UP|PFD5_ARATH (P57742) Probable prefoldin subunit 5,
partial (85%)
Length = 923
Score = 34.3 bits (77), Expect = 0.083
Identities = 20/67 (29%), Positives = 32/67 (46%), Gaps = 6/67 (8%)
Frame = +3
Query: 37 FNLPPTHPLYPDNPEPVSVQQPQ------PNPEPTTNSPHNSTTQKASEVASDATTSETP 90
F PP P P +S P+ P+P P+T+ P ++T +S ++ AT+S P
Sbjct: 324 FAPPPPASRSPPPPSTISPSAPRATTSSSPSPPPSTSPPPSTTPNTSSSTSAPATSSRKP 503
Query: 91 QHQEYST 97
+E T
Sbjct: 504 CPKEKIT 524
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 33.9 bits (76), Expect = 0.11
Identities = 36/150 (24%), Positives = 56/150 (37%), Gaps = 8/150 (5%)
Frame = +2
Query: 58 PQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHNLEKHLGGEMQPTPTKAS 117
P P P T SP AS ++ TTS +P + ST M P P ++
Sbjct: 56 PPSPPSPPTTSPACWPHTPASPPST--TTSPSPTWPKRSTAARPSPSWPSTMPPCPPSST 229
Query: 118 KTVP----EKTVLETQTETQTIPEQTVQEQTASEQVAP-DQTTSDQHIPSDQTTEQQQQP 172
T P + + T + T + P+ + + TA P + + P +T +P
Sbjct: 230 STSPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTSPTSRP 409
Query: 173 ---DSPTIIDLTSDQPSISNTTQTEPSPIP 199
SP PS SN ++ P+ P
Sbjct: 410 ARSASPPKTTTAPSTPSTSNPSRKCPTTSP 499
>TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyrate
dehydrogenase , partial (76%)
Length = 1230
Score = 33.9 bits (76), Expect = 0.11
Identities = 26/113 (23%), Positives = 48/113 (42%), Gaps = 1/113 (0%)
Frame = +2
Query: 40 PPTHPLYPDNPEPVSVQQPQPN-PEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTL 98
PP P P S P P+ P PT++SP ++T + +++ +S + +P ++
Sbjct: 179 PPPRPSPSSTSGPTSPPPPTPSQPAPTSSSPSSATPRTSAQSSSTPPPAPSPPSAPAASS 358
Query: 99 HNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAP 151
P+P+K+ +P K ++ T P T +T Q +P
Sbjct: 359 ST*PP----PNPPSPSKSRTPLPPKAATQS---TPPCPAATAARRTEP*QSSP 496
>BG043254
Length = 421
Score = 33.9 bits (76), Expect = 0.11
Identities = 19/59 (32%), Positives = 28/59 (47%)
Frame = +2
Query: 39 LPPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYST 97
L P HP P P P S P P P PT+ + S++Q A+ A++S + +T
Sbjct: 62 LRPPHPPPPPRPLPQS-HPPPPPPSPTSKTSTTSSSQSPKSSAASASSSSRSRSSAKTT 235
>AW598371 weakly similar to GP|9280319|dbj| extensin protein-like
{Arabidopsis thaliana}, partial (2%)
Length = 298
Score = 33.5 bits (75), Expect = 0.14
Identities = 14/30 (46%), Positives = 15/30 (49%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSP 69
PPTHP P P V P+P P T SP
Sbjct: 40 PPTHPRRHSPPPPTPVYSPEPGPGRTRRSP 129
Score = 28.1 bits (61), Expect = 6.0
Identities = 12/39 (30%), Positives = 18/39 (45%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKAS 78
PP P+Y P P ++ P P P P N++ A+
Sbjct: 70 PPPTPVYSPEPGPGRTRRSPPTPRPFIVVPANNSDPSAA 186
>TC220760 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (9%)
Length = 721
Score = 33.5 bits (75), Expect = 0.14
Identities = 24/100 (24%), Positives = 33/100 (33%), Gaps = 8/100 (8%)
Frame = +3
Query: 41 PTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLHN 100
P P P P P PNP P + +S+ S T S P +T +
Sbjct: 30 PPSPTSPSTPSPFPTSPAPPNPSPPRGTSPSSSATATRR*PSPTTPSALPSSTAKTTSPS 209
Query: 101 LEKHLGG--------EMQPTPTKASKTVPEKTVLETQTET 132
L G M P+P A + P + + T T
Sbjct: 210 LSSLPSGRTPGPRPHSMPPSPPPAPTSSPN*STISTPRGT 329
>TC216800 weakly similar to UP|Q9LRQ7 (Q9LRQ7) Anthocyanin 5-aromatic
acyltransferase/benzoyltransferase-like protein, partial
(24%)
Length = 2180
Score = 33.1 bits (74), Expect = 0.19
Identities = 20/49 (40%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = +1
Query: 42 THPLYPDNPEPVSVQQPQPNPEPT-TNSPHNSTTQKASEVASDATTSET 89
THP P +P P Q P +P PT T++P T+QK + TT+ET
Sbjct: 547 THPS-PSSPTPPETQSPSESPNPTQTSTPSLQTSQKLT------TTAET 672
>TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime subunit,
complete
Length = 2055
Score = 33.1 bits (74), Expect = 0.19
Identities = 29/149 (19%), Positives = 51/149 (33%), Gaps = 7/149 (4%)
Frame = +2
Query: 34 CEKFNLPPTHPLYPDNP-------EPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATT 86
CE+ +P P +P+ E +QP+P P P PH
Sbjct: 263 CEEGQIPRPRPQHPERERQQHGEKEEDEGEQPRPFPFPRPRQPHQE-------------- 400
Query: 87 SETPQHQEYSTLHNLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTAS 146
E Q +E+ EKH G + + P + + + + + +Q + S
Sbjct: 401 EEHEQKEEHEWHRKEEKHGGKGSEEEQDEREHPRPHQPHQKEEEKHEWQHKQEKHQGKES 580
Query: 147 EQVAPDQTTSDQHIPSDQTTEQQQQPDSP 175
E+ DQ ++ Q +E + P
Sbjct: 581 EEEEEDQDEDEEQDKESQESEGSESQREP 667
>TC228692 similar to UP|Q8S9A6 (Q8S9A6) Glucosyltransferase-3, partial (48%)
Length = 775
Score = 32.0 bits (71), Expect = 0.41
Identities = 36/155 (23%), Positives = 50/155 (32%), Gaps = 3/155 (1%)
Frame = +1
Query: 40 PPTHPLYPDNPEPVSVQQPQPNPEPTTNSPHNSTTQKASEVASDATTSETPQHQEYSTLH 99
PP P + P P S P PN P + P + AS + + +S
Sbjct: 199 PPPXPRHXPPPPPPSPATPPPNTSPLSPPPPPPSPSTASLRSPSSPSS------------ 342
Query: 100 NLEKHLGGEMQPTPTKASKTVPEKTVLETQTETQTIPEQTVQEQTASEQVAPDQTTSDQH 159
P P+ +S P T + T P+ + Q S AP + +
Sbjct: 343 ----------LPWPSPSSSAAPPATTSAASSATSPKPQTSKQ*SWTS*TTAPHVSPTRFK 492
Query: 160 IPSDQTTEQQQQPDSPTII---DLTSDQPSISNTT 191
P TT P SP T PS S T+
Sbjct: 493 SPLTSTT-LPAPPPSPLFFIKPSFTKPAPSPSRTS 594
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,535,698
Number of Sequences: 63676
Number of extensions: 225568
Number of successful extensions: 3660
Number of sequences better than 10.0: 236
Number of HSP's better than 10.0 without gapping: 2579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3402
length of query: 340
length of database: 12,639,632
effective HSP length: 98
effective length of query: 242
effective length of database: 6,399,384
effective search space: 1548650928
effective search space used: 1548650928
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146552.4


