

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146549.11 + phase: 0
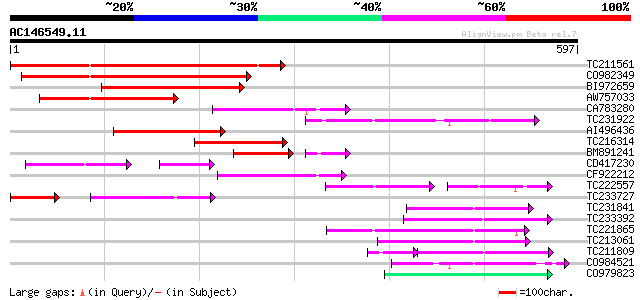
(597 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 265 3e-71
CO982349 219 4e-57
BI972659 139 4e-33
AW757033 133 3e-31
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 121 1e-27
TC231922 112 5e-25
AI496436 105 6e-23
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 100 1e-21
BM891241 84 4e-20
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 72 9e-19
CF922212 90 3e-18
TC222557 53 3e-16
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 54 3e-16
TC231841 77 2e-14
TC233392 77 3e-14
TC221865 75 1e-13
TC213061 74 2e-13
TC211809 60 3e-13
CO984521 72 7e-13
CO979823 69 8e-12
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 265 bits (678), Expect = 3e-71
Identities = 132/290 (45%), Positives = 186/290 (63%)
Frame = +1
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M +M F W KW++ECV +A++SVLVNGSPT EF QRGLRQGDPL+PFLF + AEGLN
Sbjct: 130 MWRMDFSPKWIKWIEECVKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLN 309
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
LM AV NL+ Y +G + + S LQ+ADDT+ G + NV A++ IL FE +S
Sbjct: 310 GLMRRAVEENLYKPYLVGANGVPI-SILQYADDTIFFGEAAKENVEAIKVILRSFELVSN 486
Query: 121 LKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERL 180
L++N+ KS + + W QEAA+ L C L PF+YLG+PIG ++RR WD ++ +
Sbjct: 487 LRINFAKSCFGVFGVTDQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKC 666
Query: 181 RSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSED 240
+LS+WK R++S+GGR+ L+KSVL+S+P+Y SFFR P ++ + I F WGG D
Sbjct: 667 ERKLSKWK*RHISFGGRVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPD 846
Query: 241 HRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWF 290
+KI+W+ W +C+ KE GG+ +RL + A+ C +D G F
Sbjct: 847 QKKISWIRWEKLCLPKERGGI-*KRLPNNHNAVANHNSIFCFIDFTGYTF 993
>CO982349
Length = 795
Score = 219 bits (557), Expect = 4e-57
Identities = 104/242 (42%), Positives = 157/242 (63%)
Frame = +3
Query: 13 WMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLF 72
W++ C+ +A+VS+LVN SP +EF QRGLRQGDPL+P LF + AEGL LM AV+ F
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 73 TGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVG 132
+ +G + V S LQ+ADDT+ + N++ +++IL FE SGLK+N+ +S
Sbjct: 249 NSFLVGKNKEPV-SILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGA 425
Query: 133 INIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSRLSEWKSRNL 192
I + W +EAA L+C + PF YLG+PI + R DP++ + ++L+ WK R++
Sbjct: 426 IWKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHI 605
Query: 193 SYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRKIAWVDWNTI 252
S GGR+ L+ ++L++LP+Y SFFRAP+ ++ + +I KF WGG R+IAWV W T+
Sbjct: 606 SLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWVKWETV 785
Query: 253 CM 254
C+
Sbjct: 786 CL 791
>BI972659
Length = 453
Score = 139 bits (350), Expect = 4e-33
Identities = 62/151 (41%), Positives = 98/151 (64%)
Frame = +1
Query: 97 LGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPF 156
+G SW N+ L+S+L FE +SGL++NY KS + +W +AA +L+C+ PF
Sbjct: 1 VGEASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWAHDAAQLLNCRQLDIPF 180
Query: 157 MYLGLPIGGDARRLCFWDPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFF 216
YLG+PI A W+P++ + +++LS+W + LS GGR+ L+KSVL++LP+Y LSFF
Sbjct: 181 HYLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFF 360
Query: 217 RAPAGIISSIESILIKFFWGGSEDHRKIAWV 247
+ P I+ + S+ F WGG++ H +I+WV
Sbjct: 361 KIPQRIVDKLVSLQRTFMWGGNQHHNRISWV 453
>AW757033
Length = 441
Score = 133 bits (334), Expect = 3e-31
Identities = 69/146 (47%), Positives = 94/146 (64%)
Frame = -2
Query: 32 TDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFA 91
T EF RGLRQGDPL+P LF +AAEGL LM AV N + +++G + V S LQ+A
Sbjct: 440 TREFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPV-SILQYA 264
Query: 92 DDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGINIAESWLQEAASILSCKL 151
DDT+ G + NVR +++IL FE SGLK+N+ KS I++ + W +EAA ++C L
Sbjct: 263 DDTIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFGAISVPD*WRKEAAEFMNCSL 84
Query: 152 GKTPFMYLGLPIGGDARRLCFWDPVL 177
PF YLG+PIG + RR WDP++
Sbjct: 83 LSLPFSYLGIPIGANPRRRETWDPII 6
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 121 bits (303), Expect = 1e-27
Identities = 61/148 (41%), Positives = 84/148 (56%), Gaps = 3/148 (2%)
Frame = +2
Query: 214 SFFRAPAGIISSIESILIKFFWGGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVAL 273
SFF P II+ +ES+ +F WGG D RKIAWV+W T+C+ K GGLG++ L FN L
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTL 184
Query: 274 LGKWCWRCLVDREGLWFRVLSSRYGEQGGRLKEGGMTG---SAWWREIIKIQNGVGVEGG 330
LGKW W ++ W +VL S+YG G R E G +G SAWW+++IK Q ++
Sbjct: 185 LGKWRWDLFYIQQEPWAKVLQSKYG--GWRALEEGSSGSKDSAWWKDLIKTQQ---LQRN 349
Query: 331 SWFEESISKRLGNGFNTFFWSDCWLGTE 358
+ ++G G FW D W T+
Sbjct: 350 IPLKRETIWKVGGGDRIKFWEDLWTNTD 433
>TC231922
Length = 803
Score = 112 bits (280), Expect = 5e-25
Identities = 68/257 (26%), Positives = 120/257 (46%), Gaps = 11/257 (4%)
Frame = -3
Query: 312 SAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE-SFMVRFRRLYDL 370
S WW+++ ++ N +++ R+G G FW D WL + ++ +LY +
Sbjct: 777 SHWWKDLRRLYNQPDFHS---IHQNMVWRVGCGDKIKFWQDSWLSEGCNLQQKYNQLYTI 607
Query: 371 SIHKDLSVGEMNILGWGENGEAW--RWRRRLLAWEEELVVEIRILLTNVTLQDTESDVWL 428
S +L++ +M + +N +W +WRRRL +E + V+ ++ +++Q+ D
Sbjct: 606 SRQ*NLTISKMG--KFSQNARSWEFKWRRRLFDYEYAMAVDFMNEISGISIQNQAHDSMF 433
Query: 429 WRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAP--------WHKSAPLKVSIWAWRLLRN 480
W+ + Y+ + Y++LM +S AP WH P + ++++WRL +
Sbjct: 432 WKADSSGVYSTKSAYRLLMPS-------ISPAPSRRIFQILWHLKIPPRAAVFSWRLFLD 274
Query: 481 RWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPH 540
R PT+ NL RR + D + GC Q+E L HC + LW + WI V P
Sbjct: 273 RLPTRGNLSRRSIPIQDIMCPLCGC-QHEEAGHLFFHCKMTKGLWWESMRWIRVIGALPA 97
Query: 541 QVLDHYYQFVYSSANTT 557
H+ QF A T+
Sbjct: 96 DPASHFIQFCDGFAATS 46
>AI496436
Length = 414
Score = 105 bits (262), Expect = 6e-23
Identities = 49/118 (41%), Positives = 77/118 (64%)
Frame = +1
Query: 110 SILVIFENMSGLKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARR 169
+IL FE SGLK+N+ KS I ++ W +EAA L+ + PF+YLG+PIG ++R
Sbjct: 1 TILRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLNSSVLSMPFVYLGIPIGANSRH 180
Query: 170 LCFWDPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIE 227
W+PV+ + +L+ WK + +S+GGR+ L+ SVL++LP+Y LSFFR P ++ +
Sbjct: 181 SDVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKVVDKAD 354
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 100 bits (250), Expect = 1e-21
Identities = 44/98 (44%), Positives = 62/98 (62%)
Frame = +2
Query: 195 GGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRKIAWVDWNTICM 254
GGR+ L+KSVLS+LP+ LSFF+ P I+ + ++ +F WGG++ H +I WV W IC
Sbjct: 5 GGRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICN 184
Query: 255 NKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRV 292
K GGLG++ L FN AL G+W W + LW R+
Sbjct: 185 PKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>BM891241
Length = 407
Score = 84.0 bits (206), Expect(2) = 4e-20
Identities = 31/63 (49%), Positives = 45/63 (71%)
Frame = -2
Query: 236 GGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSS 295
GG DH+KI WV W +C+ K GGLG++ + +FN+ALLGKW W D++ LW R+++S
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 296 RYG 298
+YG
Sbjct: 226 KYG 218
Score = 32.7 bits (73), Expect(2) = 4e-20
Identities = 16/47 (34%), Positives = 25/47 (53%)
Frame = -3
Query: 312 SAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE 358
S WWR++ K + S F + +S ++G G FW+D WLG +
Sbjct: 174 SYWWRDLRKFYHQ---SDHSIFHQYMSWKVGCGDKINFWTDKWLGED 43
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 72.0 bits (175), Expect(2) = 9e-19
Identities = 40/112 (35%), Positives = 61/112 (53%)
Frame = -3
Query: 17 CVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYS 76
C+ST T+S+L NG D F G+RQ DP++P+LF+L E L+ L+ +N L
Sbjct: 668 CMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQ 489
Query: 77 IGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKS 128
+ ++SHL F DD +L + V + L +F + SG KVN +K+
Sbjct: 488 VN-RGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKT 336
Score = 40.0 bits (92), Expect(2) = 9e-19
Identities = 22/58 (37%), Positives = 33/58 (55%)
Frame = -2
Query: 158 YLGLPIGGDARRLCFWDPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSF 215
YLG+PI +D +++++ R S WKS LS GRL L K V+ +LP + + F
Sbjct: 243 YLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKSNLLSMVGRLTLTK*VV*ALPSHIMQF 70
>CF922212
Length = 445
Score = 89.7 bits (221), Expect = 3e-18
Identities = 53/138 (38%), Positives = 74/138 (53%), Gaps = 2/138 (1%)
Frame = -2
Query: 219 PAGIISSIESILIKFFWGGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWC 278
P ++ + + F WGG +KIAWV+W+++C+ KE GLG R L +FN ALLGK
Sbjct: 438 PNKVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKRR 259
Query: 279 WRCLVDREGLWFRVLSSRYGEQGGRLKEGGM-TGSAWWREIIKIQNGVGVEGGSWFEE-S 336
W + L RVL S+Y +E + + S WW+EI I + E GSWFE+ +
Sbjct: 258 WNLFHHQGELGARVLDSKYKRWRNLDEERRVKSESFWWQEISFITH--STEDGSWFEKRT 85
Query: 337 ISKRLGNGFNTFFWSDCW 354
+ RLG FW D W
Sbjct: 84 KTGRLGCRAKVRFWEDGW 31
>TC222557
Length = 1002
Score = 53.1 bits (126), Expect(2) = 3e-16
Identities = 38/113 (33%), Positives = 48/113 (41%), Gaps = 3/113 (2%)
Frame = +1
Query: 462 WHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIF 521
W PLK + +AWRL++ R PTK NL RR V + C Q E L +CP
Sbjct: 442 WQLKIPLKATTFAWRLIKERIPTKGNLWRRRV-QLNNLMCPFCNRQEEEASHLFFNCPRI 618
Query: 522 GALWQQIKTW---IGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKW 571
LW + W +GVF P Q +Y Q S Q K +L W
Sbjct: 619 LPLWWESLAWTKTVGVFSPIPRQ---NYMQHTLISKGKHLQARWKCWWVALTW 768
Score = 50.4 bits (119), Expect(2) = 3e-16
Identities = 31/118 (26%), Positives = 52/118 (43%), Gaps = 3/118 (2%)
Frame = +3
Query: 333 FEESISKRLGNGFNTFFWSDCWL-GTESFMVRFRRLYDLSIHKDLSVGEMNILGWGENGE 391
F+ +I ++G G FW DCWL E ++ RLY++S + + M N
Sbjct: 51 FQSAIDWKVGCGDKIRFWEDCWLMEQEPLRAKYPRLYNISCQQQKLILVMGF--HSANVW 224
Query: 392 AWR--WRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLM 447
W+ WRR L E + ++ + D W+W+P ++ R Y+ML+
Sbjct: 225 EWKLEWRRHLFDNEVQAAASFLDDISWGHIDRWTLDCWVWKPEPNGQFSTRSAYRMLL 398
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 53.5 bits (127), Expect(2) = 3e-16
Identities = 36/133 (27%), Positives = 68/133 (51%), Gaps = 2/133 (1%)
Frame = -2
Query: 86 SHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGINIAESWLQEAAS 145
SH+ ADD ++ + NV L + ++++ G ++ +KS + +I L+ +
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLRVISD 272
Query: 146 ILSCKLGKTPFMYLGLPI--GGDARRLCFWDPVLERLRSRLSEWKSRNLSYGGRLILLKS 203
+L PF Y G+PI G RR + V ++++ +L+ WK LS GR+ L+ S
Sbjct: 271 LLQYFHSDIPFNYFGVPIFKGKPTRRHLYC--VADKIKCKLASWKGSILSIMGRIQLVNS 98
Query: 204 VLSSLPVYALSFF 216
V+ + +Y+ S +
Sbjct: 97 VIHGMLLYSFSVY 59
Score = 50.1 bits (118), Expect(2) = 3e-16
Identities = 22/52 (42%), Positives = 33/52 (63%)
Frame = -3
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLF 52
+ K G+ + W++ + +A +S+ VNG P F QRG+RQGDPLSP L+
Sbjct: 699 LNKFGYSHTFCNWIRVILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
>TC231841
Length = 791
Score = 77.0 bits (188), Expect = 2e-14
Identities = 40/134 (29%), Positives = 64/134 (46%), Gaps = 1/134 (0%)
Frame = +3
Query: 419 LQDTESDVWLWRPNIGDGYTVRGVYQMLMRQE-RHNYDVVSDAPWHKSAPLKVSIWAWRL 477
+Q + D W+W+ + YT + Y +L + D V + W P K++I+AWRL
Sbjct: 45 IQPQQGDQWVWKADPSGQYTAKSAYGVLWGEMFEEQQDGVFEELWKLKLPSKITIFAWRL 224
Query: 478 LRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFV 537
+R+R PT+ NL RR I D C E+ L HC +W + +W+ + V
Sbjct: 225 IRDRLPTRSNL-RRKQIEVDDPRCPFCRSAEESAAHLFFHCSRIAPVWWESLSWVNLLGV 401
Query: 538 DPHQVLDHYYQFVY 551
P+ H+ Q +Y
Sbjct: 402 FPNHPRQHFLQHIY 443
>TC233392
Length = 1145
Score = 76.6 bits (187), Expect = 3e-14
Identities = 45/158 (28%), Positives = 71/158 (44%), Gaps = 1/158 (0%)
Frame = -1
Query: 415 TNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQ-ERHNYDVVSDAPWHKSAPLKVSIW 473
++ +Q D W+W+ + Y+ R Y++L E + D W P K SI+
Sbjct: 686 SHAQIQQQVPDTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGALQDLWKLKIPAKASIF 507
Query: 474 AWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIG 533
AWRL+R+R PTK NL RR V+ D+ C + E + + C LW + +TW+
Sbjct: 506 AWRLIRDRLPTKSNLHRRQVVLEDS-LCPFCRIREEDASHIFLECNKIRPLWWESQTWVR 330
Query: 534 VFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKW 571
V P H+ Q V+ + + K +L W
Sbjct: 329 SLGVSPINKRQHFLQHVHGTPGSKRYNRWKTWWIALTW 216
>TC221865
Length = 1045
Score = 74.7 bits (182), Expect = 1e-13
Identities = 55/222 (24%), Positives = 97/222 (42%), Gaps = 8/222 (3%)
Frame = -3
Query: 334 EESISKRLGNGFNTFFWSDCWLGT-ESFMVRFRRLYDLSIHKDLSVGEMNILGWGENGEA 392
+ I ++G G FW D +G E+ M ++ R+Y +S + + ++ E
Sbjct: 944 QNEIVWKVGCGDKFRFWEDKLVGEGETLMRKYPRMYQISCQQQQLIQQVG--SHTETTWE 771
Query: 393 WR--WRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQ- 449
W+ WRR L E + + ++ + +D W+W+ Y+ R Y ++ +
Sbjct: 770 WKFQWRRPLFDNEVDTTIGFLEDISRFPIHRHLTDCWVWKSEPNGHYSTRSAYHLMQHEA 591
Query: 450 ERHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNE 509
+ N D + W P K +I+A RL+++R PTK+NL RR + + C+ E
Sbjct: 590 AKANTDPAFEEIWKLKIPAKAAIFA*RLVKDRLPTKNNL-RRRQVQLNGTLCLFCRNYEE 414
Query: 510 TIDLLIIHCPIFGALWQQIKTWI---GVFFVDP-HQVLDHYY 547
L C +W + +WI G F +P H L H +
Sbjct: 413 EASHLFFSCTKTQPVWWESLSWINTSGAFSANPRHHFLQHSF 288
>TC213061
Length = 823
Score = 73.6 bits (179), Expect = 2e-13
Identities = 44/164 (26%), Positives = 71/164 (42%), Gaps = 3/164 (1%)
Frame = +2
Query: 388 ENGEAWR--WRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQM 445
E G W+ WRR L E E++V + ++ D W+W Y R Y++
Sbjct: 11 ETGWEWQFQWRRPLFDSEIEMLVAFIQEVEGHKIRSDIEDQWVWAAESSGSYLARSAYRV 190
Query: 446 LMRQ-ERHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTG 504
+ D W P+KV+++AWRLL++ PT+DNL RR + C
Sbjct: 191 IREGIPEEEQDREFKELWKLKVPMKVTMFAWRLLKDILPTRDNL-RRKRVELHEYVCPLC 367
Query: 505 CGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQ 548
+E+ L HC +W + +W+ + PH H++Q
Sbjct: 368 RSMDESASHLFFHCSKILPIWWESLSWVKLVGAFPHHPRHHFHQ 499
>TC211809
Length = 895
Score = 60.5 bits (145), Expect(2) = 3e-13
Identities = 40/144 (27%), Positives = 60/144 (40%), Gaps = 1/144 (0%)
Frame = +2
Query: 430 RPNIGDGYTVRGVYQMLMRQE-RHNYDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNL 488
RP + Y+ + Y++LM D W+ P K ++AWRL+++R PT NL
Sbjct: 185 RPTLFGNYSTKSAYRLLMEATGEFPVDRTYVDLWNLKIPSKAIVFAWRLIKDRLPTWTNL 364
Query: 489 VRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQ 548
R V D++ C E L HC LW + ++W+ PH DH+ Q
Sbjct: 365 RVRQVELNDSR-CPLCNSSEEDAAHLFFHCTKTEXLWWETQSWVNSLGAFPHNPKDHFLQ 541
Query: 549 FVYSSANTTEQLLEKVKITSLKWL 572
SA K +L W+
Sbjct: 542 HDNRSAGGVXARRWKCLWVALTWV 613
Score = 33.1 bits (74), Expect(2) = 3e-13
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = +1
Query: 377 SVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWR 430
S+G GW N + WRR L E + E L VT+ DVW+W+
Sbjct: 34 SMGSQEDTGWE*N---FSWRRPLFDNEIGMAAEFLEELAQVTIHQDRPDVWVWK 186
>CO984521
Length = 716
Score = 72.0 bits (175), Expect = 7e-13
Identities = 49/193 (25%), Positives = 82/193 (42%), Gaps = 6/193 (3%)
Frame = -2
Query: 403 EEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAP- 461
E ++ V L T++ SD W W Y+ + Y+ L H+ V +
Sbjct: 712 EIDMAVAFLQQLEGFTIRPELSDQWKWAAEPSGCYSTKSAYKAL-----HHVTVGEEQDG 548
Query: 462 -----WHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLII 516
W PLKV+I+AWRL++++ PTK NL R+ + C ET L
Sbjct: 547 KFKELWKLRVPLKVAIFAWRLIQDKLPTKANL-RKKRVELQEYLCPLCRSVEETASHLFF 371
Query: 517 HCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKWLKAKN 576
HC LW + ++W+ + V P+Q H+ Q ++ ++ V + +W +
Sbjct: 370 HCSKVSPLWWESQSWVNMMGVFPYQPDQHFSQHIFGAS---------VGLQGKRW---QW 227
Query: 577 VCFPFGYHLWRQR 589
F Y +W+ R
Sbjct: 226 WWFALTYSIWKHR 188
>CO979823
Length = 853
Score = 68.6 bits (166), Expect = 8e-12
Identities = 47/178 (26%), Positives = 71/178 (39%), Gaps = 1/178 (0%)
Frame = -2
Query: 395 WRRRLLAWEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNY 454
WRR E + +T + D WLW+P G Y+ + Y +L +
Sbjct: 792 WRRPXXDSEIAMADSFLGEITQQQIHPQREDKWLWKPEPGGHYSTKSGYHVLWGELTEEI 613
Query: 455 DVVSDAP-WHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDL 513
A W P K +++AWRL+R+R PTK NL RR V+ D C E
Sbjct: 612 QDADFAEIWKLKIPTKAAVFAWRLVRDRLPTKSNLRRRQVMVQD-MVCPLCNNIEEGAAH 436
Query: 514 LIIHCPIFGALWQQIKTWIGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKW 571
L +C LW + +W+ + P H+ Q+ A+ + K +L W
Sbjct: 435 LFFNCTKTLPLWWESMSWVNLKTAMPQTPRQHFLQYGTDIADGLKSKRWKCWWIALTW 262
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,094,802
Number of Sequences: 63676
Number of extensions: 581824
Number of successful extensions: 4465
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 4289
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4410
length of query: 597
length of database: 12,639,632
effective HSP length: 103
effective length of query: 494
effective length of database: 6,081,004
effective search space: 3004015976
effective search space used: 3004015976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146549.11


