

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.3 + phase: 0
(173 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
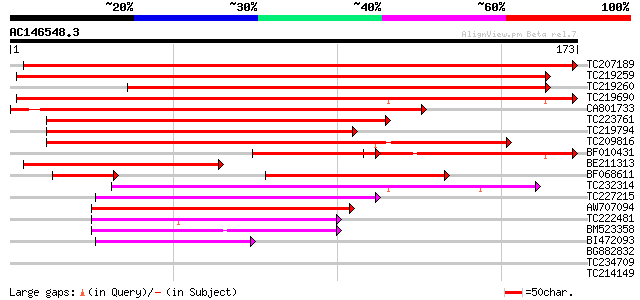
Score E
Sequences producing significant alignments: (bits) Value
TC207189 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (At... 313 2e-86
TC219259 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (At... 278 7e-76
TC219260 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (At... 223 4e-59
TC219690 similar to UP|Q9S784 (Q9S784) AtHVA22c; 50565-49239 (At... 218 1e-57
CA801733 177 2e-45
TC223761 weakly similar to UP|Q9FED2 (Q9FED2) AtHVA22e (Abscisic... 137 2e-33
TC219794 similar to UP|Q9FED2 (Q9FED2) AtHVA22e (Abscisic acid-i... 123 5e-29
TC209816 weakly similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64... 118 1e-27
BF010431 72 5e-27
BE211313 similar to PIR|C96774|C967 AtHVA22a 65476-64429 [impor... 114 3e-26
BF068611 similar to PIR|C96774|C967 AtHVA22a 65476-64429 [impor... 105 1e-23
TC232314 weakly similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial... 73 6e-14
TC227215 similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial (62%) 68 2e-12
AW707094 weakly similar to GP|21555613|gb| abscisic acid-induced... 62 1e-10
TC222481 similar to UP|Q8LEM6 (Q8LEM6) Pathogenicity protein PAT... 53 6e-08
BM523358 51 3e-07
BI472093 44 5e-05
BG882832 37 0.006
TC234709 weakly similar to UP|Q9FH69 (Q9FH69) Arabidopsis thalia... 35 0.014
TC214149 homologue to UP|PRP1_SOYBN (P08012) Repetitive proline-... 33 0.088
>TC207189 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (AtHVA22a),
partial (89%)
Length = 917
Score = 313 bits (803), Expect = 2e-86
Identities = 145/169 (85%), Positives = 159/169 (93%)
Frame = +1
Query: 5 GSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLF 64
GSGAG+FL VLL+NFDVLAGPVISLVYPLYAS+RAIESKSP+DDQQWLTYWVLYSLITLF
Sbjct: 97 GSGAGNFLKVLLKNFDVLAGPVISLVYPLYASIRAIESKSPIDDQQWLTYWVLYSLITLF 276
Query: 65 ELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKK 124
ELTFAK+LEWIPIWPYAKLI TCWLVLPYF+GAAYVYEHYVRPF NPQTINIWYVPRKK
Sbjct: 277 ELTFAKVLEWIPIWPYAKLIATCWLVLPYFSGAAYVYEHYVRPFYVNPQTINIWYVPRKK 456
Query: 125 DVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY 173
D K+DDI+TAAEKYI+ENGTEAFEN+I+RADKS+ G ++TMYDETY
Sbjct: 457 DALGKRDDILTAAEKYIQENGTEAFENIINRADKSRRGDGYYTMYDETY 603
>TC219259 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (AtHVA22a),
partial (86%)
Length = 787
Score = 278 bits (712), Expect = 7e-76
Identities = 131/163 (80%), Positives = 144/163 (87%)
Frame = +3
Query: 3 GSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLIT 62
GS GAGSFL V+L+NFDVLAGPV+SL YPLYASVRAIESKSPVDDQQWLTYWVLYSLIT
Sbjct: 72 GSNMGAGSFLKVVLKNFDVLAGPVLSLAYPLYASVRAIESKSPVDDQQWLTYWVLYSLIT 251
Query: 63 LFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPR 122
LFELTFAK+LEWIPIWPYAKLILT WLV+PYF+GAAYVYEHYVRPF N Q +NIWYVP
Sbjct: 252 LFELTFAKVLEWIPIWPYAKLILTSWLVIPYFSGAAYVYEHYVRPFFVNSQNVNIWYVPS 431
Query: 123 KKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSH 165
KKD K +D++TAAEKYIKE+GTEAFENL+ RA KS+ S H
Sbjct: 432 KKDSSGKPEDVLTAAEKYIKEHGTEAFENLLDRAGKSRKSSRH 560
>TC219260 similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429 (AtHVA22a),
partial (69%)
Length = 584
Score = 223 bits (568), Expect = 4e-59
Identities = 101/129 (78%), Positives = 114/129 (88%)
Frame = +3
Query: 37 VRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTG 96
VRAIESKSPVDDQQWLTYWVLYSLITLFELTFAK++EWIPIWPYAKLILT WLV+PYF+G
Sbjct: 3 VRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKVIEWIPIWPYAKLILTSWLVIPYFSG 182
Query: 97 AAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRA 156
AAYVYEHYVRPF NPQ +NIWYVP KKD K +D++TAAEKYI+E+GTEAFENL+ +A
Sbjct: 183 AAYVYEHYVRPFFVNPQNVNIWYVPSKKDSSGKPEDVLTAAEKYIEEHGTEAFENLLSKA 362
Query: 157 DKSKWGSSH 165
KS+ H
Sbjct: 363 GKSRKSGRH 389
>TC219690 similar to UP|Q9S784 (Q9S784) AtHVA22c; 50565-49239 (AtHVA22c),
partial (77%)
Length = 851
Score = 218 bits (554), Expect = 1e-57
Identities = 98/178 (55%), Positives = 136/178 (76%), Gaps = 7/178 (3%)
Frame = +1
Query: 3 GSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLIT 62
G+ +FL V+ NFDVLA P+++LVYPLYAS++AIE+KS DDQQWLTYW+LYS++T
Sbjct: 130 GASDNNNNFLQVVFNNFDVLALPLVTLVYPLYASIKAIETKSTTDDQQWLTYWILYSILT 309
Query: 63 LFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQT-----INI 117
+FELTFAK+LE +PIWP+AKLI +CWLVLP+F GAA VY +Y+RPF NPQ I
Sbjct: 310 IFELTFAKVLELLPIWPFAKLIFSCWLVLPHFNGAAVVYRNYIRPFYMNPQIPIPQGSQI 489
Query: 118 WYVPRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWG--SSHHTMYDETY 173
WY P+KK +F + DD+++AAE+Y++E+GTEAFE LI + D+ + ++ ++D+ Y
Sbjct: 490 WYFPQKKSLFNQPDDVLSAAERYMEEHGTEAFERLISKNDRQARARRNGNYMIFDDDY 663
>CA801733
Length = 449
Score = 177 bits (450), Expect = 2e-45
Identities = 84/127 (66%), Positives = 103/127 (80%)
Frame = +2
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSL 60
MG SG+ +FL V+ +NFDVLA P+++LVYPLYAS++AIE++S DDQQWLTYWVLYSL
Sbjct: 77 MGASGN---NFLQVVAKNFDVLALPLVTLVYPLYASIKAIETRSRTDDQQWLTYWVLYSL 247
Query: 61 ITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYV 120
ITLFELTFAK+LE + IWPYAKLIL+CWLVLP F GAA+VY HYVRPF NPQ + +
Sbjct: 248 ITLFELTFAKVLEVLAIWPYAKLILSCWLVLPNFNGAAHVYRHYVRPFYMNPQMPQMPQI 427
Query: 121 PRKKDVF 127
PR ++
Sbjct: 428 PRASQMW 448
>TC223761 weakly similar to UP|Q9FED2 (Q9FED2) AtHVA22e (Abscisic
acid-induced-like protein), partial (72%)
Length = 650
Score = 137 bits (346), Expect = 2e-33
Identities = 63/105 (60%), Positives = 78/105 (74%)
Frame = +1
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
L + R+ D + GP + L+YPLYAS+RAIES S +DDQQWLTYWVLYS +TLFEL+ KI
Sbjct: 22 LGTMARHLDTIIGPGVMLLYPLYASMRAIESPSTLDDQQWLTYWVLYSFMTLFELSTHKI 201
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTIN 116
L W PIW + KL+ WLVLP F GAAY+YE+YVR ++ N T N
Sbjct: 202 LAWFPIWVFLKLMFCVWLVLPMFNGAAYIYENYVRQYIKNIGTSN 336
>TC219794 similar to UP|Q9FED2 (Q9FED2) AtHVA22e (Abscisic acid-induced-like
protein), partial (90%)
Length = 709
Score = 123 bits (308), Expect = 5e-29
Identities = 55/95 (57%), Positives = 70/95 (72%)
Frame = +1
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
L L+ +AGPV++L+YPLYASV AIES+S +DD+QWL YW++YS +TL E+ I
Sbjct: 190 LWTLIIQLHSIAGPVVTLLYPLYASVVAIESQSKLDDEQWLAYWIIYSFLTLTEMVLQPI 369
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
LEWIPIW KL+ WLVLP F GAAY+YE +VR
Sbjct: 370 LEWIPIWYDVKLLTVAWLVLPQFAGAAYLYERFVR 474
>TC209816 weakly similar to UP|Q9S7V4 (Q9S7V4) AtHVA22a; 65476-64429
(AtHVA22a), partial (19%)
Length = 575
Score = 118 bits (296), Expect = 1e-27
Identities = 63/143 (44%), Positives = 91/143 (63%), Gaps = 1/143 (0%)
Frame = +1
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
L + L+ D LA P+++L YPL ASV+AIE+ S + + ++YW+L SLI LFE F++I
Sbjct: 1 LKLSLKCLDHLAWPLLALGYPLCASVQAIETDSNKETRDLISYWILLSLIYLFEYAFSRI 180
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLA-NPQTINIWYVPRKKDVFTKQ 130
L+W WPY KL++ LV P F A+Y Y + +R ++ NPQ I I + + F K+
Sbjct: 181 LQWFQFWPYIKLMIIFCLVTPDFGRASYAYNNLIRTCISLNPQAI-ICRLNNWRKFFVKK 357
Query: 131 DDIITAAEKYIKENGTEAFENLI 153
DD + E+Y+ ENGTEA E LI
Sbjct: 358 DDFLLHVERYLNENGTEALEKLI 426
>BF010431
Length = 391
Score = 72.0 bits (175), Expect(2) = 5e-27
Identities = 30/39 (76%), Positives = 33/39 (83%)
Frame = -1
Query: 75 IPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQ 113
+ IWPYAKLIL+CWLVLP F GAA+VY HYVRPF NPQ
Sbjct: 391 LAIWPYAKLILSCWLVLPNFNGAAHVYRHYVRPFYMNPQ 275
Score = 65.5 bits (158), Expect(2) = 5e-27
Identities = 30/67 (44%), Positives = 50/67 (73%), Gaps = 2/67 (2%)
Frame = -2
Query: 109 LANPQTINIWYVPRKKDVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWG--SSHH 166
L + + + WYVPRK ++F+KQDD++TAAE+Y++E+GTEAFE LI +AD+ + ++
Sbjct: 252 LQSQELLKWWYVPRK-NIFSKQDDVLTAAERYMEEHGTEAFERLITKADREARARRNGNY 76
Query: 167 TMYDETY 173
++D+ Y
Sbjct: 75 MIFDDDY 55
>BE211313 similar to PIR|C96774|C967 AtHVA22a 65476-64429 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 460
Score = 114 bits (284), Expect = 3e-26
Identities = 55/61 (90%), Positives = 59/61 (96%)
Frame = +3
Query: 5 GSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLF 64
GSGAG+FL VLL+NFDVLAGPVISLVYPLYAS+RAIESKSP+DDQQWLTY VLYSLITLF
Sbjct: 276 GSGAGNFLKVLLKNFDVLAGPVISLVYPLYASIRAIESKSPIDDQQWLTYCVLYSLITLF 455
Query: 65 E 65
E
Sbjct: 456 E 458
>BF068611 similar to PIR|C96774|C967 AtHVA22a 65476-64429 [imported] -
Arabidopsis thaliana, partial (42%)
Length = 246
Score = 105 bits (261), Expect = 1e-23
Identities = 44/56 (78%), Positives = 49/56 (86%)
Frame = +3
Query: 79 PYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDII 134
PYAKLI TCWLVLPYF+GAAYVYEHYVRPF NP TIN+WY+PRKKD K+DDI+
Sbjct: 78 PYAKLIATCWLVLPYFSGAAYVYEHYVRPFYVNPHTINMWYIPRKKDALGKRDDIL 245
Score = 41.2 bits (95), Expect = 2e-04
Identities = 19/20 (95%), Positives = 20/20 (100%)
Frame = +2
Query: 14 VLLRNFDVLAGPVISLVYPL 33
VLL+NFDVLAGPVISLVYPL
Sbjct: 23 VLLKNFDVLAGPVISLVYPL 82
>TC232314 weakly similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial (50%)
Length = 780
Score = 73.2 bits (178), Expect = 6e-14
Identities = 41/138 (29%), Positives = 64/138 (45%), Gaps = 7/138 (5%)
Frame = +1
Query: 32 PLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVL 91
P+Y++ +AIE+ P + Q+ L YW Y + E+ K+L WIP++ + K WL L
Sbjct: 127 PVYSTFKAIENNDPYEQQRCLLYWAAYGSFSAAEMFAEKLLSWIPLYYHMKFAFLVWLQL 306
Query: 92 PYFTGAAYVYEHYVRPFLANPQT-----INIWYVPRKKDVFTKQDDIITAAEKYIK--EN 144
P GA +Y ++RPFL Q + Y K + Q ++ A +K
Sbjct: 307 PTLDGARQLYSSHLRPFLLKHQARMDLIVEFVYGAMSKIISAYQPELEFARTLAVKFLMT 486
Query: 145 GTEAFENLIHRADKSKWG 162
T+ NLIH + G
Sbjct: 487 ATQMVRNLIHPVGRESNG 540
>TC227215 similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial (62%)
Length = 781
Score = 67.8 bits (164), Expect = 2e-12
Identities = 30/87 (34%), Positives = 48/87 (54%)
Frame = +3
Query: 27 ISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILT 86
+ + P+Y + +AIESK + L YW Y +L E+ K++ W PI+ + K
Sbjct: 156 VGVAMPVYTTFKAIESKDQDAQHKCLLYWAAYGSFSLVEVFTDKLISWCPIYYHLKFAFL 335
Query: 87 CWLVLPYFTGAAYVYEHYVRPFLANPQ 113
WL LP +GA +Y +++RPFL+ Q
Sbjct: 336 VWLQLPTTSGAKQIYANHLRPFLSRHQ 416
>AW707094 weakly similar to GP|21555613|gb| abscisic acid-induced-like
protein {Arabidopsis thaliana}, partial (33%)
Length = 262
Score = 62.4 bits (150), Expect = 1e-10
Identities = 30/80 (37%), Positives = 53/80 (65%)
Frame = +2
Query: 26 VISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLIL 85
+I+L+YPLY + I+++S ++++Q L Y ++YS +TL ++ ILE IPI KL+
Sbjct: 2 IITLLYPLYTCIITIKNQSKLNNKQELXY*IIYSFLTLTKIILQPILE*IPI*YDIKLLT 181
Query: 86 TCWLVLPYFTGAAYVYEHYV 105
L+LP FT + Y+Y+ ++
Sbjct: 182 IA*LILPQFTRSTYLYKEFM 241
>TC222481 similar to UP|Q8LEM6 (Q8LEM6) Pathogenicity protein PATH531-like
protein, partial (30%)
Length = 519
Score = 53.1 bits (126), Expect = 6e-08
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 2/78 (2%)
Frame = +3
Query: 26 VISLVYPLYASVRAIESKSPVDDQQ--WLTYWVLYSLITLFELTFAKILEWIPIWPYAKL 83
V YP Y +A+E K P +Q W YW+L +++T+ E + W+P++ AKL
Sbjct: 273 VFGYAYPAYECYKAVEKKKPEIEQLRFWCQYWILVAVLTVCERVGDTFISWVPMYXEAKL 452
Query: 84 ILTCWLVLPYFTGAAYVY 101
+L P G Y+Y
Sbjct: 453 AFFIFLWYPKTKGTTYMY 506
>BM523358
Length = 430
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/76 (30%), Positives = 41/76 (53%)
Frame = +1
Query: 26 VISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLIL 85
+ YP YA+++AIE QWL YWV++S++ + + + + ++ WIP + +
Sbjct: 190 IAGFAYPAYATLKAIEHDHDQIKSQWLIYWVIFSVLCIID-SASFLIYWIPYYYVYRFFF 366
Query: 86 TCWLVLPYFTGAAYVY 101
+L LP GA V+
Sbjct: 367 MAYLYLPISLGAKKVF 414
>BI472093
Length = 421
Score = 43.5 bits (101), Expect = 5e-05
Identities = 17/49 (34%), Positives = 28/49 (56%)
Frame = +2
Query: 27 ISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWI 75
I L P+Y++ +AIE+ P + Q+ L YW Y + E+ K+L W+
Sbjct: 113 IGLALPVYSTFKAIENNDPYEQQRCLLYWAAYGSFSAAEMFAEKLLSWV 259
>BG882832
Length = 312
Score = 36.6 bits (83), Expect = 0.006
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +1
Query: 127 FTKQDDIITAAEKYIKENGTEAFENLI 153
F K+DD + E+Y+ ENGTEA E LI
Sbjct: 34 FVKKDDFLLHVERYLNENGTEALEKLI 114
>TC234709 weakly similar to UP|Q9FH69 (Q9FH69) Arabidopsis thaliana genomic
DNA, chromosome 5, TAC clone:K16E1, partial (12%)
Length = 453
Score = 35.4 bits (80), Expect = 0.014
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Frame = +3
Query: 56 VLYSLITLFE-LTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLAN 111
V+ +L T+ E T A I W P++ KL L +L P G YVY +RP++++
Sbjct: 30 VIVALFTVLENFTDAVIGWWFPLYGELKLALFIYLWYPKTKGTGYVYNTVLRPYVSS 200
>TC214149 homologue to UP|PRP1_SOYBN (P08012) Repetitive proline-rich cell
wall protein 1 precursor, partial (96%)
Length = 942
Score = 32.7 bits (73), Expect = 0.088
Identities = 21/74 (28%), Positives = 35/74 (46%)
Frame = -1
Query: 50 QWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFL 109
QWL +L+S+ + LT KI W+ +++ WLV F V+ +V +L
Sbjct: 846 QWL*IHLLFSIYLIHPLT*FKI-RWVSSVGAWRVLWVRWLVNRRFLNRGLVHRWFVHWWL 670
Query: 110 ANPQTINIWYVPRK 123
+ +N W V R+
Sbjct: 669 LYRRLVNWWLVNRR 628
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,754,090
Number of Sequences: 63676
Number of extensions: 125696
Number of successful extensions: 724
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 721
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 173
length of database: 12,639,632
effective HSP length: 91
effective length of query: 82
effective length of database: 6,845,116
effective search space: 561299512
effective search space used: 561299512
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146548.3


